
Spinach amalgavirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Amalgaviridae; Amalgavirus
Average proteome isoelectric point is 7.72
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
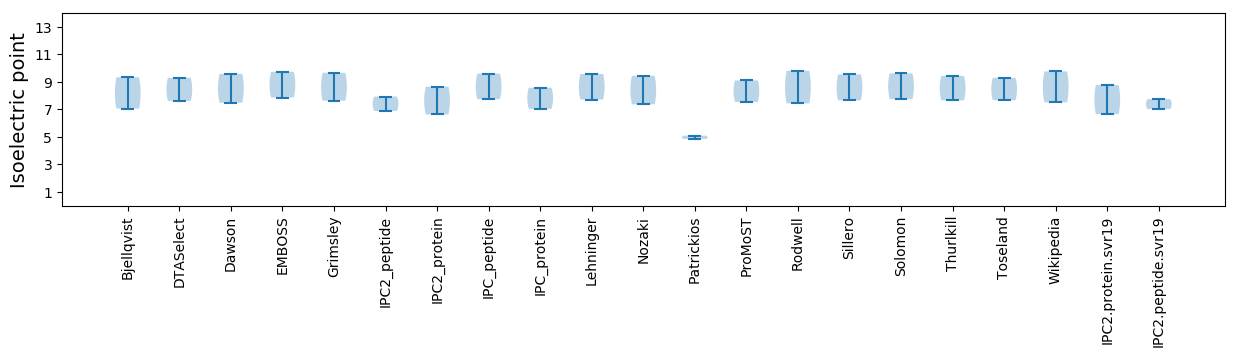
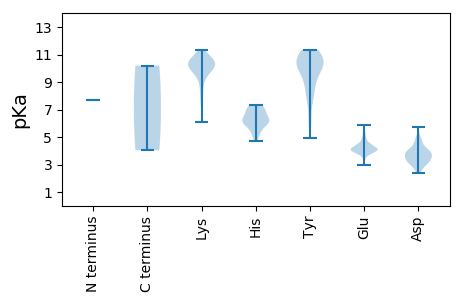
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W6S3T3|A0A1W6S3T3_9VIRU Putative coat protein OS=Spinach amalgavirus 1 OX=1985164 PE=4 SV=1
MM1 pKa = 7.7AGLEE5 pKa = 4.4GEE7 pKa = 4.43GSEE10 pKa = 5.28GIHH13 pKa = 6.25LVEE16 pKa = 4.4KK17 pKa = 10.0TPKK20 pKa = 9.84QEE22 pKa = 3.94QEE24 pKa = 4.13EE25 pKa = 4.9LFTASEE31 pKa = 4.42KK32 pKa = 10.67LRR34 pKa = 11.84EE35 pKa = 4.09AGIPLGAFDD44 pKa = 4.75RR45 pKa = 11.84NAIIKK50 pKa = 10.1AGHH53 pKa = 5.96SFNGYY58 pKa = 8.1MKK60 pKa = 9.83MIKK63 pKa = 10.25YY64 pKa = 6.31VTNYY68 pKa = 9.68TEE70 pKa = 5.41GGFFDD75 pKa = 4.06TLLVMGGSKK84 pKa = 10.31KK85 pKa = 10.51LYY87 pKa = 8.9PIHH90 pKa = 7.34SKK92 pKa = 9.41MDD94 pKa = 3.05HH95 pKa = 6.13FGFVRR100 pKa = 11.84FARR103 pKa = 11.84WLQSKK108 pKa = 9.76EE109 pKa = 4.15GQDD112 pKa = 3.97EE113 pKa = 4.86IYY115 pKa = 10.34TLQRR119 pKa = 11.84EE120 pKa = 4.39QKK122 pKa = 9.29LLRR125 pKa = 11.84KK126 pKa = 9.64AGEE129 pKa = 4.43SLTPTQMIKK138 pKa = 10.71NDD140 pKa = 3.54VFNLIRR146 pKa = 11.84TEE148 pKa = 4.18FSRR151 pKa = 11.84AMKK154 pKa = 10.53AEE156 pKa = 3.91RR157 pKa = 11.84EE158 pKa = 4.1KK159 pKa = 11.18FEE161 pKa = 4.04QEE163 pKa = 3.3KK164 pKa = 11.34DD165 pKa = 3.09EE166 pKa = 4.13LRR168 pKa = 11.84RR169 pKa = 11.84LLRR172 pKa = 11.84QKK174 pKa = 10.43EE175 pKa = 4.12RR176 pKa = 11.84EE177 pKa = 3.67EE178 pKa = 3.77RR179 pKa = 11.84QAFKK183 pKa = 11.09NLQEE187 pKa = 4.12KK188 pKa = 8.96FAPISFYY195 pKa = 10.88RR196 pKa = 11.84EE197 pKa = 3.81PTDD200 pKa = 3.47EE201 pKa = 4.17EE202 pKa = 4.37VGIAAYY208 pKa = 10.04EE209 pKa = 4.09MYY211 pKa = 10.67EE212 pKa = 3.86NEE214 pKa = 3.6ARR216 pKa = 11.84KK217 pKa = 9.86NGKK220 pKa = 7.46TPMSRR225 pKa = 11.84YY226 pKa = 9.72HH227 pKa = 6.97GGDD230 pKa = 2.89VYY232 pKa = 11.34ARR234 pKa = 11.84QHH236 pKa = 6.27FSQAARR242 pKa = 11.84EE243 pKa = 3.93LAQVYY248 pKa = 8.03FASNPEE254 pKa = 3.88NQDD257 pKa = 2.56IMEE260 pKa = 4.59RR261 pKa = 11.84FMKK264 pKa = 10.29EE265 pKa = 2.97RR266 pKa = 11.84FLFFRR271 pKa = 11.84KK272 pKa = 7.71TADD275 pKa = 3.14EE276 pKa = 4.3SGEE279 pKa = 3.98RR280 pKa = 11.84TARR283 pKa = 11.84MQLVGIGGEE292 pKa = 4.18KK293 pKa = 10.21AVEE296 pKa = 3.87DD297 pKa = 4.19ALAEE301 pKa = 4.0EE302 pKa = 4.51AKK304 pKa = 10.62AYY306 pKa = 10.6GDD308 pKa = 4.43DD309 pKa = 4.12PDD311 pKa = 5.2RR312 pKa = 11.84DD313 pKa = 3.93SAGAGAAGEE322 pKa = 4.21VQGASKK328 pKa = 10.22KK329 pKa = 10.01AKK331 pKa = 9.19PRR333 pKa = 11.84FVSKK337 pKa = 10.55PPTGRR342 pKa = 11.84GKK344 pKa = 10.27AAAGDD349 pKa = 3.97QPEE352 pKa = 4.82GGADD356 pKa = 3.44QPGSEE361 pKa = 4.45DD362 pKa = 5.7AEE364 pKa = 4.44DD365 pKa = 3.87QSDD368 pKa = 3.81TSSTLSRR375 pKa = 11.84PRR377 pKa = 11.84KK378 pKa = 6.11TARR381 pKa = 11.84NTRR384 pKa = 11.84SKK386 pKa = 10.8KK387 pKa = 10.2
MM1 pKa = 7.7AGLEE5 pKa = 4.4GEE7 pKa = 4.43GSEE10 pKa = 5.28GIHH13 pKa = 6.25LVEE16 pKa = 4.4KK17 pKa = 10.0TPKK20 pKa = 9.84QEE22 pKa = 3.94QEE24 pKa = 4.13EE25 pKa = 4.9LFTASEE31 pKa = 4.42KK32 pKa = 10.67LRR34 pKa = 11.84EE35 pKa = 4.09AGIPLGAFDD44 pKa = 4.75RR45 pKa = 11.84NAIIKK50 pKa = 10.1AGHH53 pKa = 5.96SFNGYY58 pKa = 8.1MKK60 pKa = 9.83MIKK63 pKa = 10.25YY64 pKa = 6.31VTNYY68 pKa = 9.68TEE70 pKa = 5.41GGFFDD75 pKa = 4.06TLLVMGGSKK84 pKa = 10.31KK85 pKa = 10.51LYY87 pKa = 8.9PIHH90 pKa = 7.34SKK92 pKa = 9.41MDD94 pKa = 3.05HH95 pKa = 6.13FGFVRR100 pKa = 11.84FARR103 pKa = 11.84WLQSKK108 pKa = 9.76EE109 pKa = 4.15GQDD112 pKa = 3.97EE113 pKa = 4.86IYY115 pKa = 10.34TLQRR119 pKa = 11.84EE120 pKa = 4.39QKK122 pKa = 9.29LLRR125 pKa = 11.84KK126 pKa = 9.64AGEE129 pKa = 4.43SLTPTQMIKK138 pKa = 10.71NDD140 pKa = 3.54VFNLIRR146 pKa = 11.84TEE148 pKa = 4.18FSRR151 pKa = 11.84AMKK154 pKa = 10.53AEE156 pKa = 3.91RR157 pKa = 11.84EE158 pKa = 4.1KK159 pKa = 11.18FEE161 pKa = 4.04QEE163 pKa = 3.3KK164 pKa = 11.34DD165 pKa = 3.09EE166 pKa = 4.13LRR168 pKa = 11.84RR169 pKa = 11.84LLRR172 pKa = 11.84QKK174 pKa = 10.43EE175 pKa = 4.12RR176 pKa = 11.84EE177 pKa = 3.67EE178 pKa = 3.77RR179 pKa = 11.84QAFKK183 pKa = 11.09NLQEE187 pKa = 4.12KK188 pKa = 8.96FAPISFYY195 pKa = 10.88RR196 pKa = 11.84EE197 pKa = 3.81PTDD200 pKa = 3.47EE201 pKa = 4.17EE202 pKa = 4.37VGIAAYY208 pKa = 10.04EE209 pKa = 4.09MYY211 pKa = 10.67EE212 pKa = 3.86NEE214 pKa = 3.6ARR216 pKa = 11.84KK217 pKa = 9.86NGKK220 pKa = 7.46TPMSRR225 pKa = 11.84YY226 pKa = 9.72HH227 pKa = 6.97GGDD230 pKa = 2.89VYY232 pKa = 11.34ARR234 pKa = 11.84QHH236 pKa = 6.27FSQAARR242 pKa = 11.84EE243 pKa = 3.93LAQVYY248 pKa = 8.03FASNPEE254 pKa = 3.88NQDD257 pKa = 2.56IMEE260 pKa = 4.59RR261 pKa = 11.84FMKK264 pKa = 10.29EE265 pKa = 2.97RR266 pKa = 11.84FLFFRR271 pKa = 11.84KK272 pKa = 7.71TADD275 pKa = 3.14EE276 pKa = 4.3SGEE279 pKa = 3.98RR280 pKa = 11.84TARR283 pKa = 11.84MQLVGIGGEE292 pKa = 4.18KK293 pKa = 10.21AVEE296 pKa = 3.87DD297 pKa = 4.19ALAEE301 pKa = 4.0EE302 pKa = 4.51AKK304 pKa = 10.62AYY306 pKa = 10.6GDD308 pKa = 4.43DD309 pKa = 4.12PDD311 pKa = 5.2RR312 pKa = 11.84DD313 pKa = 3.93SAGAGAAGEE322 pKa = 4.21VQGASKK328 pKa = 10.22KK329 pKa = 10.01AKK331 pKa = 9.19PRR333 pKa = 11.84FVSKK337 pKa = 10.55PPTGRR342 pKa = 11.84GKK344 pKa = 10.27AAAGDD349 pKa = 3.97QPEE352 pKa = 4.82GGADD356 pKa = 3.44QPGSEE361 pKa = 4.45DD362 pKa = 5.7AEE364 pKa = 4.44DD365 pKa = 3.87QSDD368 pKa = 3.81TSSTLSRR375 pKa = 11.84PRR377 pKa = 11.84KK378 pKa = 6.11TARR381 pKa = 11.84NTRR384 pKa = 11.84SKK386 pKa = 10.8KK387 pKa = 10.2
Molecular weight: 43.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W6S3T3|A0A1W6S3T3_9VIRU Putative coat protein OS=Spinach amalgavirus 1 OX=1985164 PE=4 SV=1
MM1 pKa = 7.7AGLEE5 pKa = 4.4GEE7 pKa = 4.43GSEE10 pKa = 5.28GIHH13 pKa = 6.25LVEE16 pKa = 4.4KK17 pKa = 10.0TPKK20 pKa = 9.84QEE22 pKa = 3.94QEE24 pKa = 4.13EE25 pKa = 4.9LFTASEE31 pKa = 4.42KK32 pKa = 10.67LRR34 pKa = 11.84EE35 pKa = 4.09AGIPLGAFDD44 pKa = 4.75RR45 pKa = 11.84NAIIKK50 pKa = 10.1AGHH53 pKa = 5.96SFNGYY58 pKa = 8.1MKK60 pKa = 9.83MIKK63 pKa = 10.25YY64 pKa = 6.31VTNYY68 pKa = 9.68TEE70 pKa = 5.41GGFFDD75 pKa = 4.06TLLVMGGSKK84 pKa = 10.31KK85 pKa = 10.51LYY87 pKa = 8.9PIHH90 pKa = 7.34SKK92 pKa = 9.41MDD94 pKa = 3.05HH95 pKa = 6.13FGFVRR100 pKa = 11.84FARR103 pKa = 11.84WLQSKK108 pKa = 9.76EE109 pKa = 4.15GQDD112 pKa = 3.97EE113 pKa = 4.86IYY115 pKa = 10.34TLQRR119 pKa = 11.84EE120 pKa = 4.39QKK122 pKa = 9.29LLRR125 pKa = 11.84KK126 pKa = 9.64AGEE129 pKa = 4.43SLTPTQMIKK138 pKa = 10.71NDD140 pKa = 3.54VFNLIRR146 pKa = 11.84TEE148 pKa = 4.18FSRR151 pKa = 11.84AMKK154 pKa = 10.53AEE156 pKa = 3.91RR157 pKa = 11.84EE158 pKa = 4.1KK159 pKa = 11.18FEE161 pKa = 4.04QEE163 pKa = 3.3KK164 pKa = 11.34DD165 pKa = 3.09EE166 pKa = 4.13LRR168 pKa = 11.84RR169 pKa = 11.84LLRR172 pKa = 11.84QKK174 pKa = 10.43EE175 pKa = 4.12RR176 pKa = 11.84EE177 pKa = 3.67EE178 pKa = 3.77RR179 pKa = 11.84QAFKK183 pKa = 11.09NLQEE187 pKa = 4.12KK188 pKa = 8.96FAPISFYY195 pKa = 10.88RR196 pKa = 11.84EE197 pKa = 3.81PTDD200 pKa = 3.47EE201 pKa = 4.17EE202 pKa = 4.37VGIAAYY208 pKa = 10.04EE209 pKa = 4.09MYY211 pKa = 10.67EE212 pKa = 3.86NEE214 pKa = 3.6ARR216 pKa = 11.84KK217 pKa = 9.86NGKK220 pKa = 7.46TPMSRR225 pKa = 11.84YY226 pKa = 9.72HH227 pKa = 6.97GGDD230 pKa = 2.89VYY232 pKa = 11.34ARR234 pKa = 11.84QHH236 pKa = 6.27FSQAARR242 pKa = 11.84EE243 pKa = 3.93LAQVYY248 pKa = 8.03FASNPEE254 pKa = 3.88NQDD257 pKa = 2.56IMEE260 pKa = 4.59RR261 pKa = 11.84FMKK264 pKa = 10.3EE265 pKa = 3.01RR266 pKa = 11.84FLFFGRR272 pKa = 11.84PPMNLGSAQQEE283 pKa = 4.61CSWLALVEE291 pKa = 4.16RR292 pKa = 11.84KK293 pKa = 8.72LWKK296 pKa = 9.69MPLQRR301 pKa = 11.84RR302 pKa = 11.84LRR304 pKa = 11.84LTGMIPIGILQAPGRR319 pKa = 11.84RR320 pKa = 11.84GRR322 pKa = 11.84CRR324 pKa = 11.84EE325 pKa = 3.98LRR327 pKa = 11.84RR328 pKa = 11.84KK329 pKa = 10.17LSPDD333 pKa = 3.26LLASPRR339 pKa = 11.84QVGVKK344 pKa = 9.43LQQGISRR351 pKa = 11.84KK352 pKa = 9.74VEE354 pKa = 3.92LTSLGPKK361 pKa = 8.44MLRR364 pKa = 11.84TSRR367 pKa = 11.84IRR369 pKa = 11.84VLRR372 pKa = 11.84SLGPEE377 pKa = 3.84RR378 pKa = 11.84PRR380 pKa = 11.84AIPVARR386 pKa = 11.84SRR388 pKa = 11.84FEE390 pKa = 3.96AGVRR394 pKa = 11.84KK395 pKa = 10.21VIGGGAMRR403 pKa = 11.84SWEE406 pKa = 4.43VDD408 pKa = 2.97SQMYY412 pKa = 10.23RR413 pKa = 11.84GGGNSADD420 pKa = 3.82ALRR423 pKa = 11.84LLGQARR429 pKa = 11.84DD430 pKa = 3.89DD431 pKa = 3.73RR432 pKa = 11.84PGAFLSGKK440 pKa = 8.03FTQASARR447 pKa = 11.84LALLLPNNLDD457 pKa = 3.5VPDD460 pKa = 4.4GAKK463 pKa = 8.35STRR466 pKa = 11.84MKK468 pKa = 10.89NFNNDD473 pKa = 2.39ATAGPFLRR481 pKa = 11.84SFGIKK486 pKa = 9.75GKK488 pKa = 10.75YY489 pKa = 7.62GLKK492 pKa = 10.15RR493 pKa = 11.84KK494 pKa = 10.1LEE496 pKa = 4.14EE497 pKa = 4.18EE498 pKa = 3.56MWRR501 pKa = 11.84YY502 pKa = 9.62YY503 pKa = 11.08DD504 pKa = 4.59DD505 pKa = 4.21YY506 pKa = 11.34GAGRR510 pKa = 11.84IDD512 pKa = 3.73SSGLPFFTARR522 pKa = 11.84VGFRR526 pKa = 11.84TKK528 pKa = 10.47LVSVEE533 pKa = 3.76KK534 pKa = 10.93AEE536 pKa = 4.48EE537 pKa = 4.14KK538 pKa = 10.74FKK540 pKa = 10.52TGQPFGRR547 pKa = 11.84AVMMLDD553 pKa = 3.64ALEE556 pKa = 4.0QAAASPLYY564 pKa = 10.4NVLSHH569 pKa = 5.37YY570 pKa = 9.11TFEE573 pKa = 4.01RR574 pKa = 11.84RR575 pKa = 11.84LRR577 pKa = 11.84RR578 pKa = 11.84DD579 pKa = 3.25CGFKK583 pKa = 10.55NAVIRR588 pKa = 11.84ASSDD592 pKa = 2.65WNEE595 pKa = 3.1IWKK598 pKa = 9.95GVRR601 pKa = 11.84EE602 pKa = 4.11AEE604 pKa = 5.06AIIEE608 pKa = 4.16LDD610 pKa = 2.75WSKK613 pKa = 10.98FDD615 pKa = 3.97RR616 pKa = 11.84EE617 pKa = 4.2RR618 pKa = 11.84PAEE621 pKa = 4.18DD622 pKa = 3.94LEE624 pKa = 4.88FIVDD628 pKa = 3.7VVISCFKK635 pKa = 10.07PRR637 pKa = 11.84NEE639 pKa = 3.86RR640 pKa = 11.84EE641 pKa = 3.75RR642 pKa = 11.84RR643 pKa = 11.84LLRR646 pKa = 11.84AYY648 pKa = 10.34GIMMRR653 pKa = 11.84RR654 pKa = 11.84ALIEE658 pKa = 4.04RR659 pKa = 11.84LLVMDD664 pKa = 4.42GGGVFGIEE672 pKa = 3.97GMVPSGSLWTGWIDD686 pKa = 3.17TALNVLYY693 pKa = 10.56LRR695 pKa = 11.84SACLEE700 pKa = 3.91VGLPSQRR707 pKa = 11.84YY708 pKa = 8.54LPMCAGDD715 pKa = 4.88DD716 pKa = 3.76NLTLFWSDD724 pKa = 3.06PGEE727 pKa = 4.2VKK729 pKa = 10.5LLKK732 pKa = 10.05IRR734 pKa = 11.84EE735 pKa = 4.45LLNEE739 pKa = 4.6WYY741 pKa = 10.28RR742 pKa = 11.84AGIDD746 pKa = 3.45DD747 pKa = 4.78ADD749 pKa = 4.24FFIHH753 pKa = 6.76RR754 pKa = 11.84PPYY757 pKa = 10.21HH758 pKa = 5.19VVKK761 pKa = 10.43RR762 pKa = 11.84QACFPPGTDD771 pKa = 3.32LKK773 pKa = 10.97GGTSMLMKK781 pKa = 10.1DD782 pKa = 3.8AEE784 pKa = 4.23WVEE787 pKa = 4.25FEE789 pKa = 4.08GEE791 pKa = 4.16LRR793 pKa = 11.84VDD795 pKa = 3.79EE796 pKa = 4.49AAGRR800 pKa = 11.84SHH802 pKa = 7.02RR803 pKa = 11.84WQYY806 pKa = 10.95LFKK809 pKa = 10.77GRR811 pKa = 11.84PKK813 pKa = 10.46FLSCYY818 pKa = 4.9WTRR821 pKa = 11.84DD822 pKa = 3.37GLPIRR827 pKa = 11.84PTSDD831 pKa = 3.16NLEE834 pKa = 4.07KK835 pKa = 10.94LLWPEE840 pKa = 5.26GIHH843 pKa = 7.21KK844 pKa = 10.88SIDD847 pKa = 3.44DD848 pKa = 3.97YY849 pKa = 10.88EE850 pKa = 5.84ASVASMAVDD859 pKa = 3.42NPFNHH864 pKa = 6.72HH865 pKa = 6.04NVNHH869 pKa = 5.88MLMRR873 pKa = 11.84YY874 pKa = 9.69VIIQQIRR881 pKa = 11.84RR882 pKa = 11.84VSAGILSPEE891 pKa = 3.98EE892 pKa = 3.81CLAFCKK898 pKa = 10.2FRR900 pKa = 11.84SQEE903 pKa = 4.08GEE905 pKa = 4.12EE906 pKa = 4.03IPYY909 pKa = 10.62PMVAPWRR916 pKa = 11.84RR917 pKa = 11.84GMHH920 pKa = 5.5EE921 pKa = 4.78ARR923 pKa = 11.84MEE925 pKa = 4.71DD926 pKa = 3.84YY927 pKa = 10.88PEE929 pKa = 4.01TKK931 pKa = 9.62PWVKK935 pKa = 10.21EE936 pKa = 3.62FRR938 pKa = 11.84DD939 pKa = 4.82FISGVSSLYY948 pKa = 10.43ARR950 pKa = 11.84KK951 pKa = 8.27PTGGVDD957 pKa = 3.51AYY959 pKa = 11.16RR960 pKa = 11.84FMEE963 pKa = 4.95IIRR966 pKa = 11.84GEE968 pKa = 3.96QSPGEE973 pKa = 4.12GQFGSDD979 pKa = 3.12LVKK982 pKa = 10.35WVDD985 pKa = 3.39WLRR988 pKa = 11.84KK989 pKa = 9.53HH990 pKa = 6.28PVTKK994 pKa = 10.56FLRR997 pKa = 11.84ATGGRR1002 pKa = 11.84HH1003 pKa = 4.68HH1004 pKa = 6.68QKK1006 pKa = 8.83STARR1010 pKa = 11.84LEE1012 pKa = 4.57GEE1014 pKa = 4.39DD1015 pKa = 3.6LRR1017 pKa = 11.84KK1018 pKa = 10.37VEE1020 pKa = 4.4DD1021 pKa = 3.21AFMALRR1027 pKa = 11.84EE1028 pKa = 4.14RR1029 pKa = 11.84LGSGQVGSTEE1039 pKa = 5.05DD1040 pKa = 3.47FSLWVSDD1047 pKa = 3.9LLRR1050 pKa = 11.84SGMM1053 pKa = 4.04
MM1 pKa = 7.7AGLEE5 pKa = 4.4GEE7 pKa = 4.43GSEE10 pKa = 5.28GIHH13 pKa = 6.25LVEE16 pKa = 4.4KK17 pKa = 10.0TPKK20 pKa = 9.84QEE22 pKa = 3.94QEE24 pKa = 4.13EE25 pKa = 4.9LFTASEE31 pKa = 4.42KK32 pKa = 10.67LRR34 pKa = 11.84EE35 pKa = 4.09AGIPLGAFDD44 pKa = 4.75RR45 pKa = 11.84NAIIKK50 pKa = 10.1AGHH53 pKa = 5.96SFNGYY58 pKa = 8.1MKK60 pKa = 9.83MIKK63 pKa = 10.25YY64 pKa = 6.31VTNYY68 pKa = 9.68TEE70 pKa = 5.41GGFFDD75 pKa = 4.06TLLVMGGSKK84 pKa = 10.31KK85 pKa = 10.51LYY87 pKa = 8.9PIHH90 pKa = 7.34SKK92 pKa = 9.41MDD94 pKa = 3.05HH95 pKa = 6.13FGFVRR100 pKa = 11.84FARR103 pKa = 11.84WLQSKK108 pKa = 9.76EE109 pKa = 4.15GQDD112 pKa = 3.97EE113 pKa = 4.86IYY115 pKa = 10.34TLQRR119 pKa = 11.84EE120 pKa = 4.39QKK122 pKa = 9.29LLRR125 pKa = 11.84KK126 pKa = 9.64AGEE129 pKa = 4.43SLTPTQMIKK138 pKa = 10.71NDD140 pKa = 3.54VFNLIRR146 pKa = 11.84TEE148 pKa = 4.18FSRR151 pKa = 11.84AMKK154 pKa = 10.53AEE156 pKa = 3.91RR157 pKa = 11.84EE158 pKa = 4.1KK159 pKa = 11.18FEE161 pKa = 4.04QEE163 pKa = 3.3KK164 pKa = 11.34DD165 pKa = 3.09EE166 pKa = 4.13LRR168 pKa = 11.84RR169 pKa = 11.84LLRR172 pKa = 11.84QKK174 pKa = 10.43EE175 pKa = 4.12RR176 pKa = 11.84EE177 pKa = 3.67EE178 pKa = 3.77RR179 pKa = 11.84QAFKK183 pKa = 11.09NLQEE187 pKa = 4.12KK188 pKa = 8.96FAPISFYY195 pKa = 10.88RR196 pKa = 11.84EE197 pKa = 3.81PTDD200 pKa = 3.47EE201 pKa = 4.17EE202 pKa = 4.37VGIAAYY208 pKa = 10.04EE209 pKa = 4.09MYY211 pKa = 10.67EE212 pKa = 3.86NEE214 pKa = 3.6ARR216 pKa = 11.84KK217 pKa = 9.86NGKK220 pKa = 7.46TPMSRR225 pKa = 11.84YY226 pKa = 9.72HH227 pKa = 6.97GGDD230 pKa = 2.89VYY232 pKa = 11.34ARR234 pKa = 11.84QHH236 pKa = 6.27FSQAARR242 pKa = 11.84EE243 pKa = 3.93LAQVYY248 pKa = 8.03FASNPEE254 pKa = 3.88NQDD257 pKa = 2.56IMEE260 pKa = 4.59RR261 pKa = 11.84FMKK264 pKa = 10.3EE265 pKa = 3.01RR266 pKa = 11.84FLFFGRR272 pKa = 11.84PPMNLGSAQQEE283 pKa = 4.61CSWLALVEE291 pKa = 4.16RR292 pKa = 11.84KK293 pKa = 8.72LWKK296 pKa = 9.69MPLQRR301 pKa = 11.84RR302 pKa = 11.84LRR304 pKa = 11.84LTGMIPIGILQAPGRR319 pKa = 11.84RR320 pKa = 11.84GRR322 pKa = 11.84CRR324 pKa = 11.84EE325 pKa = 3.98LRR327 pKa = 11.84RR328 pKa = 11.84KK329 pKa = 10.17LSPDD333 pKa = 3.26LLASPRR339 pKa = 11.84QVGVKK344 pKa = 9.43LQQGISRR351 pKa = 11.84KK352 pKa = 9.74VEE354 pKa = 3.92LTSLGPKK361 pKa = 8.44MLRR364 pKa = 11.84TSRR367 pKa = 11.84IRR369 pKa = 11.84VLRR372 pKa = 11.84SLGPEE377 pKa = 3.84RR378 pKa = 11.84PRR380 pKa = 11.84AIPVARR386 pKa = 11.84SRR388 pKa = 11.84FEE390 pKa = 3.96AGVRR394 pKa = 11.84KK395 pKa = 10.21VIGGGAMRR403 pKa = 11.84SWEE406 pKa = 4.43VDD408 pKa = 2.97SQMYY412 pKa = 10.23RR413 pKa = 11.84GGGNSADD420 pKa = 3.82ALRR423 pKa = 11.84LLGQARR429 pKa = 11.84DD430 pKa = 3.89DD431 pKa = 3.73RR432 pKa = 11.84PGAFLSGKK440 pKa = 8.03FTQASARR447 pKa = 11.84LALLLPNNLDD457 pKa = 3.5VPDD460 pKa = 4.4GAKK463 pKa = 8.35STRR466 pKa = 11.84MKK468 pKa = 10.89NFNNDD473 pKa = 2.39ATAGPFLRR481 pKa = 11.84SFGIKK486 pKa = 9.75GKK488 pKa = 10.75YY489 pKa = 7.62GLKK492 pKa = 10.15RR493 pKa = 11.84KK494 pKa = 10.1LEE496 pKa = 4.14EE497 pKa = 4.18EE498 pKa = 3.56MWRR501 pKa = 11.84YY502 pKa = 9.62YY503 pKa = 11.08DD504 pKa = 4.59DD505 pKa = 4.21YY506 pKa = 11.34GAGRR510 pKa = 11.84IDD512 pKa = 3.73SSGLPFFTARR522 pKa = 11.84VGFRR526 pKa = 11.84TKK528 pKa = 10.47LVSVEE533 pKa = 3.76KK534 pKa = 10.93AEE536 pKa = 4.48EE537 pKa = 4.14KK538 pKa = 10.74FKK540 pKa = 10.52TGQPFGRR547 pKa = 11.84AVMMLDD553 pKa = 3.64ALEE556 pKa = 4.0QAAASPLYY564 pKa = 10.4NVLSHH569 pKa = 5.37YY570 pKa = 9.11TFEE573 pKa = 4.01RR574 pKa = 11.84RR575 pKa = 11.84LRR577 pKa = 11.84RR578 pKa = 11.84DD579 pKa = 3.25CGFKK583 pKa = 10.55NAVIRR588 pKa = 11.84ASSDD592 pKa = 2.65WNEE595 pKa = 3.1IWKK598 pKa = 9.95GVRR601 pKa = 11.84EE602 pKa = 4.11AEE604 pKa = 5.06AIIEE608 pKa = 4.16LDD610 pKa = 2.75WSKK613 pKa = 10.98FDD615 pKa = 3.97RR616 pKa = 11.84EE617 pKa = 4.2RR618 pKa = 11.84PAEE621 pKa = 4.18DD622 pKa = 3.94LEE624 pKa = 4.88FIVDD628 pKa = 3.7VVISCFKK635 pKa = 10.07PRR637 pKa = 11.84NEE639 pKa = 3.86RR640 pKa = 11.84EE641 pKa = 3.75RR642 pKa = 11.84RR643 pKa = 11.84LLRR646 pKa = 11.84AYY648 pKa = 10.34GIMMRR653 pKa = 11.84RR654 pKa = 11.84ALIEE658 pKa = 4.04RR659 pKa = 11.84LLVMDD664 pKa = 4.42GGGVFGIEE672 pKa = 3.97GMVPSGSLWTGWIDD686 pKa = 3.17TALNVLYY693 pKa = 10.56LRR695 pKa = 11.84SACLEE700 pKa = 3.91VGLPSQRR707 pKa = 11.84YY708 pKa = 8.54LPMCAGDD715 pKa = 4.88DD716 pKa = 3.76NLTLFWSDD724 pKa = 3.06PGEE727 pKa = 4.2VKK729 pKa = 10.5LLKK732 pKa = 10.05IRR734 pKa = 11.84EE735 pKa = 4.45LLNEE739 pKa = 4.6WYY741 pKa = 10.28RR742 pKa = 11.84AGIDD746 pKa = 3.45DD747 pKa = 4.78ADD749 pKa = 4.24FFIHH753 pKa = 6.76RR754 pKa = 11.84PPYY757 pKa = 10.21HH758 pKa = 5.19VVKK761 pKa = 10.43RR762 pKa = 11.84QACFPPGTDD771 pKa = 3.32LKK773 pKa = 10.97GGTSMLMKK781 pKa = 10.1DD782 pKa = 3.8AEE784 pKa = 4.23WVEE787 pKa = 4.25FEE789 pKa = 4.08GEE791 pKa = 4.16LRR793 pKa = 11.84VDD795 pKa = 3.79EE796 pKa = 4.49AAGRR800 pKa = 11.84SHH802 pKa = 7.02RR803 pKa = 11.84WQYY806 pKa = 10.95LFKK809 pKa = 10.77GRR811 pKa = 11.84PKK813 pKa = 10.46FLSCYY818 pKa = 4.9WTRR821 pKa = 11.84DD822 pKa = 3.37GLPIRR827 pKa = 11.84PTSDD831 pKa = 3.16NLEE834 pKa = 4.07KK835 pKa = 10.94LLWPEE840 pKa = 5.26GIHH843 pKa = 7.21KK844 pKa = 10.88SIDD847 pKa = 3.44DD848 pKa = 3.97YY849 pKa = 10.88EE850 pKa = 5.84ASVASMAVDD859 pKa = 3.42NPFNHH864 pKa = 6.72HH865 pKa = 6.04NVNHH869 pKa = 5.88MLMRR873 pKa = 11.84YY874 pKa = 9.69VIIQQIRR881 pKa = 11.84RR882 pKa = 11.84VSAGILSPEE891 pKa = 3.98EE892 pKa = 3.81CLAFCKK898 pKa = 10.2FRR900 pKa = 11.84SQEE903 pKa = 4.08GEE905 pKa = 4.12EE906 pKa = 4.03IPYY909 pKa = 10.62PMVAPWRR916 pKa = 11.84RR917 pKa = 11.84GMHH920 pKa = 5.5EE921 pKa = 4.78ARR923 pKa = 11.84MEE925 pKa = 4.71DD926 pKa = 3.84YY927 pKa = 10.88PEE929 pKa = 4.01TKK931 pKa = 9.62PWVKK935 pKa = 10.21EE936 pKa = 3.62FRR938 pKa = 11.84DD939 pKa = 4.82FISGVSSLYY948 pKa = 10.43ARR950 pKa = 11.84KK951 pKa = 8.27PTGGVDD957 pKa = 3.51AYY959 pKa = 11.16RR960 pKa = 11.84FMEE963 pKa = 4.95IIRR966 pKa = 11.84GEE968 pKa = 3.96QSPGEE973 pKa = 4.12GQFGSDD979 pKa = 3.12LVKK982 pKa = 10.35WVDD985 pKa = 3.39WLRR988 pKa = 11.84KK989 pKa = 9.53HH990 pKa = 6.28PVTKK994 pKa = 10.56FLRR997 pKa = 11.84ATGGRR1002 pKa = 11.84HH1003 pKa = 4.68HH1004 pKa = 6.68QKK1006 pKa = 8.83STARR1010 pKa = 11.84LEE1012 pKa = 4.57GEE1014 pKa = 4.39DD1015 pKa = 3.6LRR1017 pKa = 11.84KK1018 pKa = 10.37VEE1020 pKa = 4.4DD1021 pKa = 3.21AFMALRR1027 pKa = 11.84EE1028 pKa = 4.14RR1029 pKa = 11.84LGSGQVGSTEE1039 pKa = 5.05DD1040 pKa = 3.47FSLWVSDD1047 pKa = 3.9LLRR1050 pKa = 11.84SGMM1053 pKa = 4.04
Molecular weight: 120.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1440 |
387 |
1053 |
720.0 |
82.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.917 ± 1.141 | 0.694 ± 0.367 |
5.0 ± 0.089 | 9.236 ± 1.126 |
5.208 ± 0.115 | 8.542 ± 0.129 |
1.667 ± 0.061 | 4.028 ± 0.353 |
6.597 ± 1.019 | 8.542 ± 1.372 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.403 ± 0.159 | 2.778 ± 0.034 |
4.514 ± 0.337 | 3.958 ± 0.639 |
9.375 ± 0.72 | 5.903 ± 0.115 |
3.611 ± 0.549 | 4.514 ± 0.746 |
1.528 ± 0.67 | 2.986 ± 0.061 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
