
Oscillibacter sp. CAG:155
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Oscillibacter; environmental samples
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
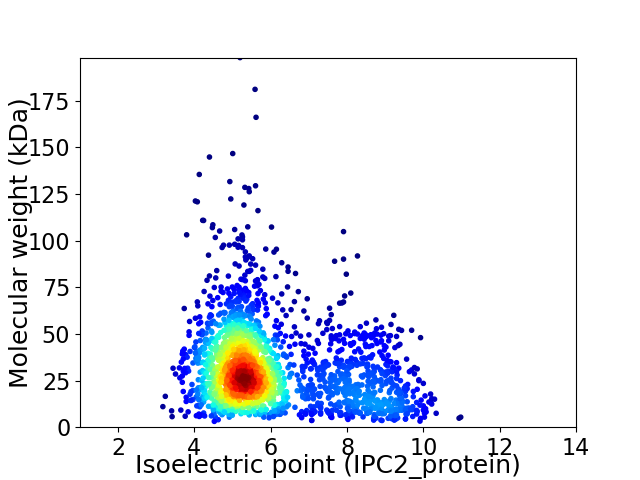
Virtual 2D-PAGE plot for 2148 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
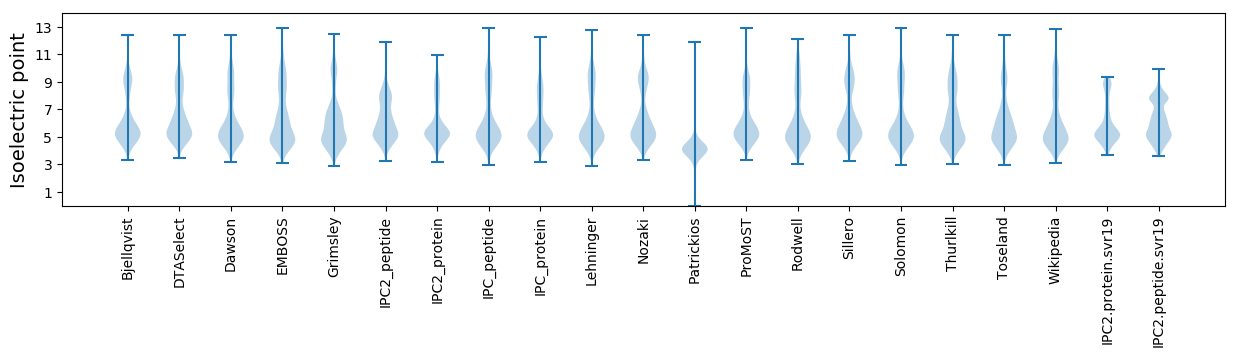
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6UAW7|R6UAW7_9FIRM HTH cro/C1-type domain-containing protein OS=Oscillibacter sp. CAG:155 OX=1262910 GN=BN503_00927 PE=4 SV=1
MM1 pKa = 7.43KK2 pKa = 10.38KK3 pKa = 10.2KK4 pKa = 10.55LLSLVMAAAMVLGLAACGSNSDD26 pKa = 3.72TADD29 pKa = 3.14TSGSDD34 pKa = 3.73DD35 pKa = 3.8SASGTTFKK43 pKa = 10.59IGTIGPLTGEE53 pKa = 3.81NAIYY57 pKa = 10.57GLAVANGAKK66 pKa = 9.9IAVEE70 pKa = 4.56EE71 pKa = 4.43INASDD76 pKa = 3.68SAIKK80 pKa = 10.59FEE82 pKa = 4.94LQSQDD87 pKa = 3.43DD88 pKa = 4.14VADD91 pKa = 4.27GEE93 pKa = 4.83TSVNAYY99 pKa = 9.81NKK101 pKa = 10.51LMDD104 pKa = 3.44WGMQLLVGPTTTGAAQAVSAVTNTDD129 pKa = 2.91RR130 pKa = 11.84TFMLTPSASSTDD142 pKa = 3.8VIDD145 pKa = 4.73GKK147 pKa = 10.99DD148 pKa = 3.27NVFQVCFTDD157 pKa = 4.09PNQGSSSADD166 pKa = 3.12YY167 pKa = 9.31MAEE170 pKa = 3.84NMPDD174 pKa = 3.11AKK176 pKa = 10.42IAIIYY181 pKa = 10.25RR182 pKa = 11.84NDD184 pKa = 3.28DD185 pKa = 3.68AYY187 pKa = 11.59SQGIHH192 pKa = 6.11DD193 pKa = 4.31TFVTEE198 pKa = 4.68AAAKK202 pKa = 9.04GLNVVYY208 pKa = 10.32EE209 pKa = 4.41GTFTKK214 pKa = 9.93DD215 pKa = 2.43TATDD219 pKa = 3.9FSVQLTGAQSAGADD233 pKa = 3.57LVFLPIYY240 pKa = 7.46YY241 pKa = 10.04QPASVILNQANAMGYY256 pKa = 9.92APTFFGVDD264 pKa = 3.29GMDD267 pKa = 5.2GILTMEE273 pKa = 4.66GFDD276 pKa = 3.28TSLAEE281 pKa = 4.23GVMLLTPFSATLEE294 pKa = 4.25EE295 pKa = 4.25NQSFVQAYY303 pKa = 8.89QDD305 pKa = 3.68EE306 pKa = 4.97FGDD309 pKa = 4.0TPNQFAADD317 pKa = 4.08AYY319 pKa = 10.35DD320 pKa = 3.21AVYY323 pKa = 10.23ILKK326 pKa = 10.32AALEE330 pKa = 4.29AAEE333 pKa = 4.81CTPDD337 pKa = 3.41MSSADD342 pKa = 3.22ICEE345 pKa = 3.9ALIPVMPTLSYY356 pKa = 11.35DD357 pKa = 3.4GLTGKK362 pKa = 10.7GMTWDD367 pKa = 3.17ASGAVSKK374 pKa = 11.11DD375 pKa = 2.84PTAFVIEE382 pKa = 4.28NGTYY386 pKa = 10.39VLPP389 pKa = 4.73
MM1 pKa = 7.43KK2 pKa = 10.38KK3 pKa = 10.2KK4 pKa = 10.55LLSLVMAAAMVLGLAACGSNSDD26 pKa = 3.72TADD29 pKa = 3.14TSGSDD34 pKa = 3.73DD35 pKa = 3.8SASGTTFKK43 pKa = 10.59IGTIGPLTGEE53 pKa = 3.81NAIYY57 pKa = 10.57GLAVANGAKK66 pKa = 9.9IAVEE70 pKa = 4.56EE71 pKa = 4.43INASDD76 pKa = 3.68SAIKK80 pKa = 10.59FEE82 pKa = 4.94LQSQDD87 pKa = 3.43DD88 pKa = 4.14VADD91 pKa = 4.27GEE93 pKa = 4.83TSVNAYY99 pKa = 9.81NKK101 pKa = 10.51LMDD104 pKa = 3.44WGMQLLVGPTTTGAAQAVSAVTNTDD129 pKa = 2.91RR130 pKa = 11.84TFMLTPSASSTDD142 pKa = 3.8VIDD145 pKa = 4.73GKK147 pKa = 10.99DD148 pKa = 3.27NVFQVCFTDD157 pKa = 4.09PNQGSSSADD166 pKa = 3.12YY167 pKa = 9.31MAEE170 pKa = 3.84NMPDD174 pKa = 3.11AKK176 pKa = 10.42IAIIYY181 pKa = 10.25RR182 pKa = 11.84NDD184 pKa = 3.28DD185 pKa = 3.68AYY187 pKa = 11.59SQGIHH192 pKa = 6.11DD193 pKa = 4.31TFVTEE198 pKa = 4.68AAAKK202 pKa = 9.04GLNVVYY208 pKa = 10.32EE209 pKa = 4.41GTFTKK214 pKa = 9.93DD215 pKa = 2.43TATDD219 pKa = 3.9FSVQLTGAQSAGADD233 pKa = 3.57LVFLPIYY240 pKa = 7.46YY241 pKa = 10.04QPASVILNQANAMGYY256 pKa = 9.92APTFFGVDD264 pKa = 3.29GMDD267 pKa = 5.2GILTMEE273 pKa = 4.66GFDD276 pKa = 3.28TSLAEE281 pKa = 4.23GVMLLTPFSATLEE294 pKa = 4.25EE295 pKa = 4.25NQSFVQAYY303 pKa = 8.89QDD305 pKa = 3.68EE306 pKa = 4.97FGDD309 pKa = 4.0TPNQFAADD317 pKa = 4.08AYY319 pKa = 10.35DD320 pKa = 3.21AVYY323 pKa = 10.23ILKK326 pKa = 10.32AALEE330 pKa = 4.29AAEE333 pKa = 4.81CTPDD337 pKa = 3.41MSSADD342 pKa = 3.22ICEE345 pKa = 3.9ALIPVMPTLSYY356 pKa = 11.35DD357 pKa = 3.4GLTGKK362 pKa = 10.7GMTWDD367 pKa = 3.17ASGAVSKK374 pKa = 11.11DD375 pKa = 2.84PTAFVIEE382 pKa = 4.28NGTYY386 pKa = 10.39VLPP389 pKa = 4.73
Molecular weight: 40.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6UBB9|R6UBB9_9FIRM ABC transporter permease protein OS=Oscillibacter sp. CAG:155 OX=1262910 GN=BN503_01035 PE=3 SV=1
MM1 pKa = 7.29GAFVKK6 pKa = 10.35KK7 pKa = 10.02IHH9 pKa = 7.05LSTLLSNGLWSLLGLALLLWPAPSARR35 pKa = 11.84AVCLALGAVLLLCGAGNLAAALPNRR60 pKa = 11.84DD61 pKa = 3.29EE62 pKa = 4.41TLYY65 pKa = 10.17TALRR69 pKa = 11.84LGSGVILAVIGLWLLFRR86 pKa = 11.84PLLAAALIPKK96 pKa = 8.55LTGILLCIHH105 pKa = 6.96GGSSLGDD112 pKa = 3.2AMLLRR117 pKa = 11.84RR118 pKa = 11.84GGDD121 pKa = 3.63ARR123 pKa = 11.84WSAAVLGGAISLALGLLLFFLAVPAFLIAARR154 pKa = 11.84IVGACLLWDD163 pKa = 4.13GVSGLWIASRR173 pKa = 11.84VGHH176 pKa = 7.12LPRR179 pKa = 11.84QDD181 pKa = 3.1PSGRR185 pKa = 3.58
MM1 pKa = 7.29GAFVKK6 pKa = 10.35KK7 pKa = 10.02IHH9 pKa = 7.05LSTLLSNGLWSLLGLALLLWPAPSARR35 pKa = 11.84AVCLALGAVLLLCGAGNLAAALPNRR60 pKa = 11.84DD61 pKa = 3.29EE62 pKa = 4.41TLYY65 pKa = 10.17TALRR69 pKa = 11.84LGSGVILAVIGLWLLFRR86 pKa = 11.84PLLAAALIPKK96 pKa = 8.55LTGILLCIHH105 pKa = 6.96GGSSLGDD112 pKa = 3.2AMLLRR117 pKa = 11.84RR118 pKa = 11.84GGDD121 pKa = 3.63ARR123 pKa = 11.84WSAAVLGGAISLALGLLLFFLAVPAFLIAARR154 pKa = 11.84IVGACLLWDD163 pKa = 4.13GVSGLWIASRR173 pKa = 11.84VGHH176 pKa = 7.12LPRR179 pKa = 11.84QDD181 pKa = 3.1PSGRR185 pKa = 3.58
Molecular weight: 19.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
634855 |
29 |
1836 |
295.6 |
32.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.915 ± 0.055 | 1.711 ± 0.024 |
5.592 ± 0.044 | 6.689 ± 0.048 |
3.658 ± 0.033 | 8.156 ± 0.053 |
1.929 ± 0.028 | 5.388 ± 0.042 |
4.324 ± 0.053 | 10.219 ± 0.074 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.741 ± 0.027 | 3.012 ± 0.033 |
4.23 ± 0.034 | 3.632 ± 0.041 |
5.739 ± 0.057 | 5.442 ± 0.038 |
5.802 ± 0.042 | 7.527 ± 0.057 |
1.023 ± 0.021 | 3.267 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
