
Avon-Heathcote Estuary associated circular virus 13
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.23
Get precalculated fractions of proteins
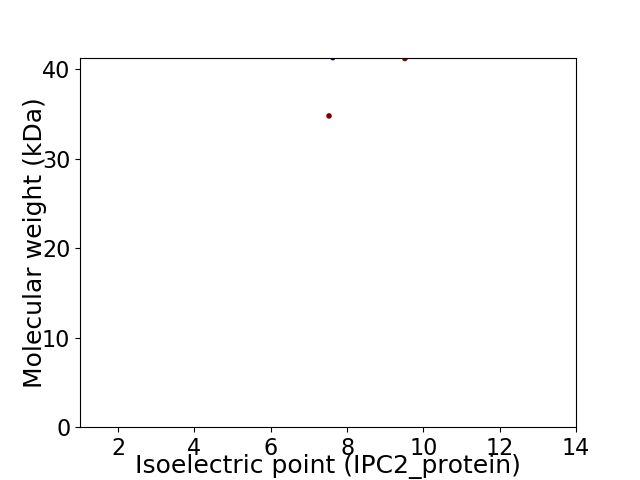
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IB23|A0A0C5IB23_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 13 OX=1618236 PE=3 SV=1
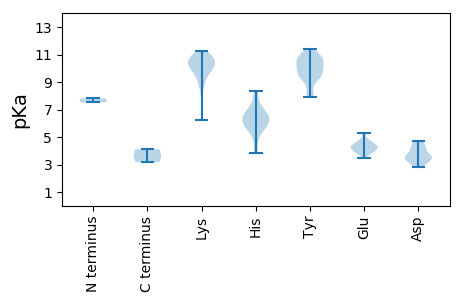
MM1 pKa = 7.52TCRR4 pKa = 11.84NWCLTLNNFTPEE16 pKa = 3.87EE17 pKa = 4.12EE18 pKa = 4.21EE19 pKa = 4.09VLKK22 pKa = 10.91EE23 pKa = 3.84EE24 pKa = 4.32KK25 pKa = 9.91PWIKK29 pKa = 10.1FVVYY33 pKa = 9.22QVEE36 pKa = 4.43MGEE39 pKa = 4.12SGTPHH44 pKa = 5.61LQGYY48 pKa = 9.5IEE50 pKa = 4.51TNGGVRR56 pKa = 11.84MSKK59 pKa = 8.9MKK61 pKa = 10.64RR62 pKa = 11.84MLEE65 pKa = 4.02RR66 pKa = 11.84GHH68 pKa = 6.17WEE70 pKa = 3.45QRR72 pKa = 11.84LGSRR76 pKa = 11.84AQALTYY82 pKa = 10.08CLKK85 pKa = 10.09EE86 pKa = 4.2DD87 pKa = 3.48SRR89 pKa = 11.84VRR91 pKa = 11.84GPYY94 pKa = 10.41LIGLSQEE101 pKa = 4.2EE102 pKa = 4.38LNSYY106 pKa = 9.51LKK108 pKa = 10.67KK109 pKa = 10.56NPKK112 pKa = 8.85STDD115 pKa = 3.22LNVIKK120 pKa = 10.69DD121 pKa = 3.87KK122 pKa = 11.04IKK124 pKa = 10.98AGASEE129 pKa = 5.29LDD131 pKa = 3.39VADD134 pKa = 3.54AHH136 pKa = 6.21FGSWVRR142 pKa = 11.84YY143 pKa = 7.95YY144 pKa = 10.98KK145 pKa = 10.61AFEE148 pKa = 4.31RR149 pKa = 11.84YY150 pKa = 9.04RR151 pKa = 11.84CLNTKK156 pKa = 9.61KK157 pKa = 10.38RR158 pKa = 11.84STIDD162 pKa = 3.0KK163 pKa = 10.09VIIIVGPTGTGKK175 pKa = 10.38SRR177 pKa = 11.84FCMEE181 pKa = 4.71QYY183 pKa = 10.85PDD185 pKa = 4.6AYY187 pKa = 9.38WKK189 pKa = 10.07QRR191 pKa = 11.84SNWWCGYY198 pKa = 9.49SGEE201 pKa = 4.29STIIIDD207 pKa = 4.7EE208 pKa = 4.71FYY210 pKa = 11.05GWIPFDD216 pKa = 3.5MLLRR220 pKa = 11.84LCDD223 pKa = 4.2RR224 pKa = 11.84YY225 pKa = 10.39PLLLEE230 pKa = 4.37TKK232 pKa = 10.18GGQCQCVATTIVITSNHH249 pKa = 6.39HH250 pKa = 5.7PNTWYY255 pKa = 10.41KK256 pKa = 10.17NSYY259 pKa = 9.69VEE261 pKa = 3.78SLYY264 pKa = 10.91RR265 pKa = 11.84RR266 pKa = 11.84FTNIIYY272 pKa = 9.25MPALGVSEE280 pKa = 4.57HH281 pKa = 6.61LNSIEE286 pKa = 4.68DD287 pKa = 3.32LHH289 pKa = 8.32LNYY292 pKa = 10.26FNFPPNN298 pKa = 3.17
MM1 pKa = 7.52TCRR4 pKa = 11.84NWCLTLNNFTPEE16 pKa = 3.87EE17 pKa = 4.12EE18 pKa = 4.21EE19 pKa = 4.09VLKK22 pKa = 10.91EE23 pKa = 3.84EE24 pKa = 4.32KK25 pKa = 9.91PWIKK29 pKa = 10.1FVVYY33 pKa = 9.22QVEE36 pKa = 4.43MGEE39 pKa = 4.12SGTPHH44 pKa = 5.61LQGYY48 pKa = 9.5IEE50 pKa = 4.51TNGGVRR56 pKa = 11.84MSKK59 pKa = 8.9MKK61 pKa = 10.64RR62 pKa = 11.84MLEE65 pKa = 4.02RR66 pKa = 11.84GHH68 pKa = 6.17WEE70 pKa = 3.45QRR72 pKa = 11.84LGSRR76 pKa = 11.84AQALTYY82 pKa = 10.08CLKK85 pKa = 10.09EE86 pKa = 4.2DD87 pKa = 3.48SRR89 pKa = 11.84VRR91 pKa = 11.84GPYY94 pKa = 10.41LIGLSQEE101 pKa = 4.2EE102 pKa = 4.38LNSYY106 pKa = 9.51LKK108 pKa = 10.67KK109 pKa = 10.56NPKK112 pKa = 8.85STDD115 pKa = 3.22LNVIKK120 pKa = 10.69DD121 pKa = 3.87KK122 pKa = 11.04IKK124 pKa = 10.98AGASEE129 pKa = 5.29LDD131 pKa = 3.39VADD134 pKa = 3.54AHH136 pKa = 6.21FGSWVRR142 pKa = 11.84YY143 pKa = 7.95YY144 pKa = 10.98KK145 pKa = 10.61AFEE148 pKa = 4.31RR149 pKa = 11.84YY150 pKa = 9.04RR151 pKa = 11.84CLNTKK156 pKa = 9.61KK157 pKa = 10.38RR158 pKa = 11.84STIDD162 pKa = 3.0KK163 pKa = 10.09VIIIVGPTGTGKK175 pKa = 10.38SRR177 pKa = 11.84FCMEE181 pKa = 4.71QYY183 pKa = 10.85PDD185 pKa = 4.6AYY187 pKa = 9.38WKK189 pKa = 10.07QRR191 pKa = 11.84SNWWCGYY198 pKa = 9.49SGEE201 pKa = 4.29STIIIDD207 pKa = 4.7EE208 pKa = 4.71FYY210 pKa = 11.05GWIPFDD216 pKa = 3.5MLLRR220 pKa = 11.84LCDD223 pKa = 4.2RR224 pKa = 11.84YY225 pKa = 10.39PLLLEE230 pKa = 4.37TKK232 pKa = 10.18GGQCQCVATTIVITSNHH249 pKa = 6.39HH250 pKa = 5.7PNTWYY255 pKa = 10.41KK256 pKa = 10.17NSYY259 pKa = 9.69VEE261 pKa = 3.78SLYY264 pKa = 10.91RR265 pKa = 11.84RR266 pKa = 11.84FTNIIYY272 pKa = 9.25MPALGVSEE280 pKa = 4.57HH281 pKa = 6.61LNSIEE286 pKa = 4.68DD287 pKa = 3.32LHH289 pKa = 8.32LNYY292 pKa = 10.26FNFPPNN298 pKa = 3.17
Molecular weight: 34.8 kDa
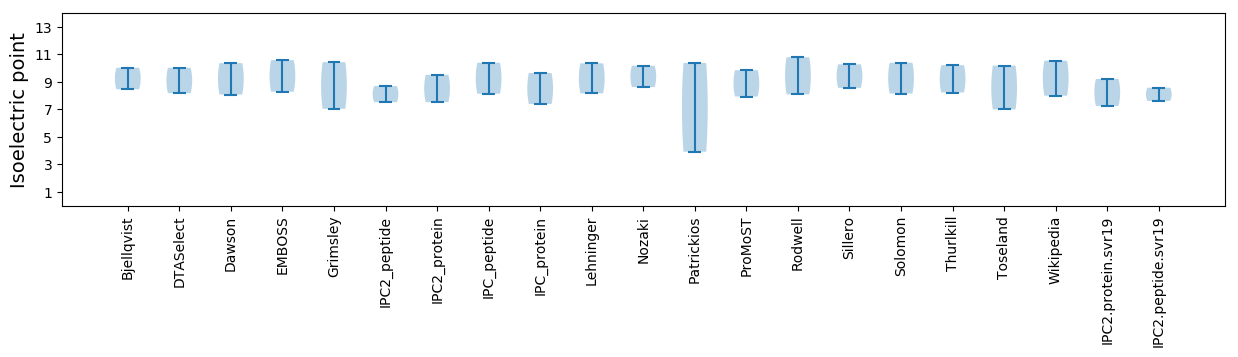
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IB23|A0A0C5IB23_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 13 OX=1618236 PE=3 SV=1
MM1 pKa = 7.81RR2 pKa = 11.84LLKK5 pKa = 10.37PKK7 pKa = 9.92EE8 pKa = 3.75YY9 pKa = 10.5SQGKK13 pKa = 9.75RR14 pKa = 11.84IFGQAFATAGAAAMRR29 pKa = 11.84ARR31 pKa = 11.84KK32 pKa = 9.46RR33 pKa = 11.84SRR35 pKa = 11.84GYY37 pKa = 10.69SRR39 pKa = 11.84TNTGRR44 pKa = 11.84RR45 pKa = 11.84TGMRR49 pKa = 11.84SKK51 pKa = 10.79KK52 pKa = 10.2RR53 pKa = 11.84IGLGRR58 pKa = 11.84TLTTRR63 pKa = 11.84QKK65 pKa = 10.9DD66 pKa = 3.76FSRR69 pKa = 11.84TYY71 pKa = 9.68VRR73 pKa = 11.84KK74 pKa = 9.65PMPYY78 pKa = 8.13RR79 pKa = 11.84KK80 pKa = 9.31KK81 pKa = 10.64RR82 pKa = 11.84KK83 pKa = 8.2WVQFKK88 pKa = 11.28NKK90 pKa = 9.29VNTVAEE96 pKa = 4.72DD97 pKa = 3.63EE98 pKa = 5.29LGTQTVIFNNQDD110 pKa = 2.97TAGITEE116 pKa = 4.73GVAGGQGTLEE126 pKa = 3.92ASLYY130 pKa = 10.13GVAGRR135 pKa = 11.84PDD137 pKa = 3.39RR138 pKa = 11.84HH139 pKa = 6.34NDD141 pKa = 3.19LAIVRR146 pKa = 11.84GMLNDD151 pKa = 4.17GDD153 pKa = 3.94PTAAAGINVDD163 pKa = 4.2DD164 pKa = 3.63TTKK167 pKa = 10.89YY168 pKa = 9.71IFHH171 pKa = 6.9SGVLDD176 pKa = 3.09ITLRR180 pKa = 11.84NTSKK184 pKa = 10.37QRR186 pKa = 11.84EE187 pKa = 4.37TANDD191 pKa = 4.11PYY193 pKa = 10.84IPWNEE198 pKa = 3.97CSLEE202 pKa = 3.82VDD204 pKa = 4.55LYY206 pKa = 10.76IISSNKK212 pKa = 8.68KK213 pKa = 7.16WQPDD217 pKa = 3.58TLSIRR222 pKa = 11.84QCLDD226 pKa = 2.9SGFATTKK233 pKa = 10.83GLDD236 pKa = 3.43GAGNQLTRR244 pKa = 11.84SKK246 pKa = 10.84RR247 pKa = 11.84GVTVFDD253 pKa = 4.52CPDD256 pKa = 4.51GISKK260 pKa = 10.9YY261 pKa = 10.6GLQVQKK267 pKa = 9.33KK268 pKa = 6.23TKK270 pKa = 10.0YY271 pKa = 9.8FIPYY275 pKa = 8.01GQQVTYY281 pKa = 9.71QMRR284 pKa = 11.84DD285 pKa = 3.44PKK287 pKa = 10.17RR288 pKa = 11.84HH289 pKa = 3.83VMRR292 pKa = 11.84RR293 pKa = 11.84GNMSTGNYY301 pKa = 9.6GNTPNKK307 pKa = 9.83PGLTHH312 pKa = 6.32YY313 pKa = 10.8VFISYY318 pKa = 10.33KK319 pKa = 10.21PVTGLPNLADD329 pKa = 3.63INNIPQLSMGNTRR342 pKa = 11.84KK343 pKa = 10.32YY344 pKa = 7.99MFKK347 pKa = 10.68VEE349 pKa = 4.18GMNDD353 pKa = 2.82TRR355 pKa = 11.84DD356 pKa = 3.66EE357 pKa = 4.35YY358 pKa = 11.44KK359 pKa = 10.64SGSIVDD365 pKa = 4.17FNN367 pKa = 4.11
MM1 pKa = 7.81RR2 pKa = 11.84LLKK5 pKa = 10.37PKK7 pKa = 9.92EE8 pKa = 3.75YY9 pKa = 10.5SQGKK13 pKa = 9.75RR14 pKa = 11.84IFGQAFATAGAAAMRR29 pKa = 11.84ARR31 pKa = 11.84KK32 pKa = 9.46RR33 pKa = 11.84SRR35 pKa = 11.84GYY37 pKa = 10.69SRR39 pKa = 11.84TNTGRR44 pKa = 11.84RR45 pKa = 11.84TGMRR49 pKa = 11.84SKK51 pKa = 10.79KK52 pKa = 10.2RR53 pKa = 11.84IGLGRR58 pKa = 11.84TLTTRR63 pKa = 11.84QKK65 pKa = 10.9DD66 pKa = 3.76FSRR69 pKa = 11.84TYY71 pKa = 9.68VRR73 pKa = 11.84KK74 pKa = 9.65PMPYY78 pKa = 8.13RR79 pKa = 11.84KK80 pKa = 9.31KK81 pKa = 10.64RR82 pKa = 11.84KK83 pKa = 8.2WVQFKK88 pKa = 11.28NKK90 pKa = 9.29VNTVAEE96 pKa = 4.72DD97 pKa = 3.63EE98 pKa = 5.29LGTQTVIFNNQDD110 pKa = 2.97TAGITEE116 pKa = 4.73GVAGGQGTLEE126 pKa = 3.92ASLYY130 pKa = 10.13GVAGRR135 pKa = 11.84PDD137 pKa = 3.39RR138 pKa = 11.84HH139 pKa = 6.34NDD141 pKa = 3.19LAIVRR146 pKa = 11.84GMLNDD151 pKa = 4.17GDD153 pKa = 3.94PTAAAGINVDD163 pKa = 4.2DD164 pKa = 3.63TTKK167 pKa = 10.89YY168 pKa = 9.71IFHH171 pKa = 6.9SGVLDD176 pKa = 3.09ITLRR180 pKa = 11.84NTSKK184 pKa = 10.37QRR186 pKa = 11.84EE187 pKa = 4.37TANDD191 pKa = 4.11PYY193 pKa = 10.84IPWNEE198 pKa = 3.97CSLEE202 pKa = 3.82VDD204 pKa = 4.55LYY206 pKa = 10.76IISSNKK212 pKa = 8.68KK213 pKa = 7.16WQPDD217 pKa = 3.58TLSIRR222 pKa = 11.84QCLDD226 pKa = 2.9SGFATTKK233 pKa = 10.83GLDD236 pKa = 3.43GAGNQLTRR244 pKa = 11.84SKK246 pKa = 10.84RR247 pKa = 11.84GVTVFDD253 pKa = 4.52CPDD256 pKa = 4.51GISKK260 pKa = 10.9YY261 pKa = 10.6GLQVQKK267 pKa = 9.33KK268 pKa = 6.23TKK270 pKa = 10.0YY271 pKa = 9.8FIPYY275 pKa = 8.01GQQVTYY281 pKa = 9.71QMRR284 pKa = 11.84DD285 pKa = 3.44PKK287 pKa = 10.17RR288 pKa = 11.84HH289 pKa = 3.83VMRR292 pKa = 11.84RR293 pKa = 11.84GNMSTGNYY301 pKa = 9.6GNTPNKK307 pKa = 9.83PGLTHH312 pKa = 6.32YY313 pKa = 10.8VFISYY318 pKa = 10.33KK319 pKa = 10.21PVTGLPNLADD329 pKa = 3.63INNIPQLSMGNTRR342 pKa = 11.84KK343 pKa = 10.32YY344 pKa = 7.99MFKK347 pKa = 10.68VEE349 pKa = 4.18GMNDD353 pKa = 2.82TRR355 pKa = 11.84DD356 pKa = 3.66EE357 pKa = 4.35YY358 pKa = 11.44KK359 pKa = 10.64SGSIVDD365 pKa = 4.17FNN367 pKa = 4.11
Molecular weight: 41.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
665 |
298 |
367 |
332.5 |
38.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.511 ± 0.759 | 1.805 ± 0.798 |
4.962 ± 0.834 | 4.962 ± 1.809 |
3.308 ± 0.031 | 8.421 ± 1.122 |
1.654 ± 0.456 | 5.414 ± 0.411 |
7.368 ± 0.431 | 7.218 ± 0.989 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.857 ± 0.113 | 6.015 ± 0.204 |
4.361 ± 0.001 | 3.91 ± 0.584 |
7.368 ± 0.872 | 5.865 ± 0.336 |
7.669 ± 1.069 | 5.263 ± 0.151 |
1.805 ± 0.798 | 5.263 ± 0.51 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
