
Tenacibaculum sp. M341
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Tenacibaculum; unclassified Tenacibaculum
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
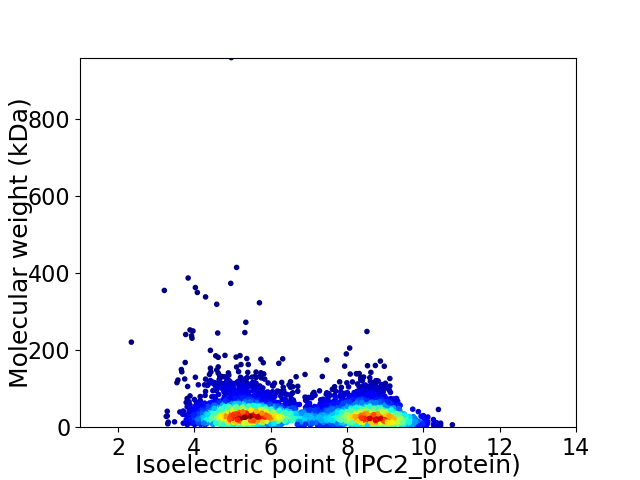
Virtual 2D-PAGE plot for 4253 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R1A672|A0A4R1A672_9FLAO Uncharacterized protein OS=Tenacibaculum sp. M341 OX=2530339 GN=EYW44_15235 PE=4 SV=1
MM1 pKa = 7.64KK2 pKa = 9.06KK3 pKa = 8.21TLLFLIYY10 pKa = 10.35FSLILFANAQDD21 pKa = 4.05GASTCDD27 pKa = 3.29AAEE30 pKa = 4.27PMCSDD35 pKa = 3.86DD36 pKa = 3.76MGVKK40 pKa = 10.0IFDD43 pKa = 3.86NVTGKK48 pKa = 9.98PNTGTIACLSTTPNPAWFFIRR69 pKa = 11.84VEE71 pKa = 3.93SSGRR75 pKa = 11.84LEE77 pKa = 3.89FQIIQAQDD85 pKa = 2.55FDD87 pKa = 3.96AAGNVVGNTLDD98 pKa = 3.42VDD100 pKa = 4.64FTSWGPFTAPDD111 pKa = 4.29GNCSSLATTCVDD123 pKa = 3.23TTGAPTTCVDD133 pKa = 3.15NTFDD137 pKa = 3.84PDD139 pKa = 3.84YY140 pKa = 10.86YY141 pKa = 10.49INNEE145 pKa = 4.18DD146 pKa = 3.53GTNIMDD152 pKa = 4.24CSYY155 pKa = 10.68SGSAIEE161 pKa = 5.58SFTIEE166 pKa = 3.87NAVAGEE172 pKa = 4.04FYY174 pKa = 10.63ILLITNFQDD183 pKa = 3.58SPGKK187 pKa = 10.24IKK189 pKa = 10.94LEE191 pKa = 3.88QTNLGTTGAGEE202 pKa = 4.31TDD204 pKa = 3.58CSIIAGEE211 pKa = 4.66LGPDD215 pKa = 3.16QTVCDD220 pKa = 4.26GTTITLDD227 pKa = 3.52GTPSTGTATSYY238 pKa = 10.07EE239 pKa = 4.07WQIDD243 pKa = 3.45TGAGFTTIAGEE254 pKa = 4.24TGNTLTINNDD264 pKa = 2.51KK265 pKa = 10.95SGTYY269 pKa = 9.62RR270 pKa = 11.84VIITDD275 pKa = 3.8DD276 pKa = 3.5VGGTATDD283 pKa = 5.05DD284 pKa = 3.19IDD286 pKa = 3.68ITFLPVPVANKK297 pKa = 9.28PADD300 pKa = 3.5INFCDD305 pKa = 3.8SDD307 pKa = 4.0SDD309 pKa = 3.99GFHH312 pKa = 6.61TFDD315 pKa = 4.36LQNDD319 pKa = 3.69VTPLVLGAQSAIQFEE334 pKa = 4.75VVYY337 pKa = 10.15FLSQTDD343 pKa = 3.29ADD345 pKa = 4.33NNNTANALANPYY357 pKa = 9.36TNSTANSTQTIYY369 pKa = 11.41ARR371 pKa = 11.84IHH373 pKa = 5.22NTSNPDD379 pKa = 2.96TCFDD383 pKa = 3.66TTSFSLDD390 pKa = 3.2INSLPTPRR398 pKa = 11.84QPSDD402 pKa = 2.99IVVCNDD408 pKa = 3.29LGNPVGFYY416 pKa = 11.23NNFILATKK424 pKa = 10.29DD425 pKa = 3.39NEE427 pKa = 4.24ILGGLSPTQYY437 pKa = 10.25SVSYY441 pKa = 8.43HH442 pKa = 4.65TTLAGAQTNATTDD455 pKa = 3.68VIDD458 pKa = 4.63KK459 pKa = 10.73NIAYY463 pKa = 8.86TNIIPNEE470 pKa = 3.64QVIFVRR476 pKa = 11.84VEE478 pKa = 3.71NSSNTSCSVATTVSSATFKK497 pKa = 10.64PFKK500 pKa = 10.52LVVNPLPVIINPIVQIEE517 pKa = 4.07QCEE520 pKa = 3.99NDD522 pKa = 3.42GDD524 pKa = 3.92FTAIINLTQAQINISNNHH542 pKa = 5.61LNEE545 pKa = 3.78TFKK548 pKa = 11.29YY549 pKa = 7.77YY550 pKa = 10.36TSEE553 pKa = 3.73VDD555 pKa = 3.81ANTDD559 pKa = 3.34NAPIADD565 pKa = 5.05PINYY569 pKa = 8.23NATNGEE575 pKa = 4.58VVWVRR580 pKa = 11.84TISDD584 pKa = 3.48KK585 pKa = 11.5DD586 pKa = 3.68CFRR589 pKa = 11.84VSQINVIISFAADD602 pKa = 2.69IMYY605 pKa = 10.47DD606 pKa = 3.92KK607 pKa = 11.07EE608 pKa = 4.14FTEE611 pKa = 5.7CDD613 pKa = 3.61DD614 pKa = 5.46FLDD617 pKa = 4.51ADD619 pKa = 4.31GNNTPSNDD627 pKa = 3.54DD628 pKa = 3.19TDD630 pKa = 5.56GITNFDD636 pKa = 3.42FSIAEE641 pKa = 4.05GEE643 pKa = 4.27IINLFDD649 pKa = 3.61PAIRR653 pKa = 11.84SDD655 pKa = 3.63LKK657 pKa = 11.07VLFFEE662 pKa = 4.65TQADD666 pKa = 3.76RR667 pKa = 11.84DD668 pKa = 3.84AVTNEE673 pKa = 3.36IADD676 pKa = 3.29ISKK679 pKa = 10.24YY680 pKa = 10.66RR681 pKa = 11.84NTNFPGLASNTIYY694 pKa = 10.55IKK696 pKa = 10.43IINKK700 pKa = 9.41INNNCTGLSKK710 pKa = 10.92LFVRR714 pKa = 11.84VLPLPSFDD722 pKa = 3.43VTSPQILCLNNLSSIEE738 pKa = 4.04AEE740 pKa = 4.5SPGDD744 pKa = 3.37TYY746 pKa = 11.42AYY748 pKa = 9.07EE749 pKa = 3.88WTRR752 pKa = 11.84NGNATVIGNNQTLNLTQGGEE772 pKa = 4.34YY773 pKa = 10.26KK774 pKa = 9.46VTAINTTTLCKK785 pKa = 9.62RR786 pKa = 11.84SRR788 pKa = 11.84IIFVNEE794 pKa = 3.93SIIATINQNDD804 pKa = 3.51VSIIDD809 pKa = 4.64DD810 pKa = 3.88SDD812 pKa = 4.04NNTITINNDD821 pKa = 2.84TGNLGIGAYY830 pKa = 9.42EE831 pKa = 4.52FALQDD836 pKa = 3.33EE837 pKa = 4.92NGQIVQDD844 pKa = 4.09YY845 pKa = 9.8QDD847 pKa = 3.45EE848 pKa = 4.58PKK850 pKa = 10.66FEE852 pKa = 4.07NLQGGIYY859 pKa = 9.58TILVRR864 pKa = 11.84DD865 pKa = 4.09KK866 pKa = 10.02NNCGIAEE873 pKa = 4.48LDD875 pKa = 3.65VSVLEE880 pKa = 4.7FPDD883 pKa = 4.53FFTPNEE889 pKa = 3.96DD890 pKa = 4.96SINDD894 pKa = 3.36TWNVKK899 pKa = 8.94GANSLFYY906 pKa = 9.56PSNSIYY912 pKa = 10.61IFDD915 pKa = 4.41RR916 pKa = 11.84YY917 pKa = 10.64GKK919 pKa = 10.32LLTSLKK925 pKa = 10.46VDD927 pKa = 3.49DD928 pKa = 4.45QGWNGTYY935 pKa = 10.36NGNALPSNDD944 pKa = 2.8YY945 pKa = 9.62WFNIEE950 pKa = 3.88LTDD953 pKa = 4.13RR954 pKa = 11.84NGNVITRR961 pKa = 11.84KK962 pKa = 9.6GHH964 pKa = 6.3FSLLLDD970 pKa = 3.79
MM1 pKa = 7.64KK2 pKa = 9.06KK3 pKa = 8.21TLLFLIYY10 pKa = 10.35FSLILFANAQDD21 pKa = 4.05GASTCDD27 pKa = 3.29AAEE30 pKa = 4.27PMCSDD35 pKa = 3.86DD36 pKa = 3.76MGVKK40 pKa = 10.0IFDD43 pKa = 3.86NVTGKK48 pKa = 9.98PNTGTIACLSTTPNPAWFFIRR69 pKa = 11.84VEE71 pKa = 3.93SSGRR75 pKa = 11.84LEE77 pKa = 3.89FQIIQAQDD85 pKa = 2.55FDD87 pKa = 3.96AAGNVVGNTLDD98 pKa = 3.42VDD100 pKa = 4.64FTSWGPFTAPDD111 pKa = 4.29GNCSSLATTCVDD123 pKa = 3.23TTGAPTTCVDD133 pKa = 3.15NTFDD137 pKa = 3.84PDD139 pKa = 3.84YY140 pKa = 10.86YY141 pKa = 10.49INNEE145 pKa = 4.18DD146 pKa = 3.53GTNIMDD152 pKa = 4.24CSYY155 pKa = 10.68SGSAIEE161 pKa = 5.58SFTIEE166 pKa = 3.87NAVAGEE172 pKa = 4.04FYY174 pKa = 10.63ILLITNFQDD183 pKa = 3.58SPGKK187 pKa = 10.24IKK189 pKa = 10.94LEE191 pKa = 3.88QTNLGTTGAGEE202 pKa = 4.31TDD204 pKa = 3.58CSIIAGEE211 pKa = 4.66LGPDD215 pKa = 3.16QTVCDD220 pKa = 4.26GTTITLDD227 pKa = 3.52GTPSTGTATSYY238 pKa = 10.07EE239 pKa = 4.07WQIDD243 pKa = 3.45TGAGFTTIAGEE254 pKa = 4.24TGNTLTINNDD264 pKa = 2.51KK265 pKa = 10.95SGTYY269 pKa = 9.62RR270 pKa = 11.84VIITDD275 pKa = 3.8DD276 pKa = 3.5VGGTATDD283 pKa = 5.05DD284 pKa = 3.19IDD286 pKa = 3.68ITFLPVPVANKK297 pKa = 9.28PADD300 pKa = 3.5INFCDD305 pKa = 3.8SDD307 pKa = 4.0SDD309 pKa = 3.99GFHH312 pKa = 6.61TFDD315 pKa = 4.36LQNDD319 pKa = 3.69VTPLVLGAQSAIQFEE334 pKa = 4.75VVYY337 pKa = 10.15FLSQTDD343 pKa = 3.29ADD345 pKa = 4.33NNNTANALANPYY357 pKa = 9.36TNSTANSTQTIYY369 pKa = 11.41ARR371 pKa = 11.84IHH373 pKa = 5.22NTSNPDD379 pKa = 2.96TCFDD383 pKa = 3.66TTSFSLDD390 pKa = 3.2INSLPTPRR398 pKa = 11.84QPSDD402 pKa = 2.99IVVCNDD408 pKa = 3.29LGNPVGFYY416 pKa = 11.23NNFILATKK424 pKa = 10.29DD425 pKa = 3.39NEE427 pKa = 4.24ILGGLSPTQYY437 pKa = 10.25SVSYY441 pKa = 8.43HH442 pKa = 4.65TTLAGAQTNATTDD455 pKa = 3.68VIDD458 pKa = 4.63KK459 pKa = 10.73NIAYY463 pKa = 8.86TNIIPNEE470 pKa = 3.64QVIFVRR476 pKa = 11.84VEE478 pKa = 3.71NSSNTSCSVATTVSSATFKK497 pKa = 10.64PFKK500 pKa = 10.52LVVNPLPVIINPIVQIEE517 pKa = 4.07QCEE520 pKa = 3.99NDD522 pKa = 3.42GDD524 pKa = 3.92FTAIINLTQAQINISNNHH542 pKa = 5.61LNEE545 pKa = 3.78TFKK548 pKa = 11.29YY549 pKa = 7.77YY550 pKa = 10.36TSEE553 pKa = 3.73VDD555 pKa = 3.81ANTDD559 pKa = 3.34NAPIADD565 pKa = 5.05PINYY569 pKa = 8.23NATNGEE575 pKa = 4.58VVWVRR580 pKa = 11.84TISDD584 pKa = 3.48KK585 pKa = 11.5DD586 pKa = 3.68CFRR589 pKa = 11.84VSQINVIISFAADD602 pKa = 2.69IMYY605 pKa = 10.47DD606 pKa = 3.92KK607 pKa = 11.07EE608 pKa = 4.14FTEE611 pKa = 5.7CDD613 pKa = 3.61DD614 pKa = 5.46FLDD617 pKa = 4.51ADD619 pKa = 4.31GNNTPSNDD627 pKa = 3.54DD628 pKa = 3.19TDD630 pKa = 5.56GITNFDD636 pKa = 3.42FSIAEE641 pKa = 4.05GEE643 pKa = 4.27IINLFDD649 pKa = 3.61PAIRR653 pKa = 11.84SDD655 pKa = 3.63LKK657 pKa = 11.07VLFFEE662 pKa = 4.65TQADD666 pKa = 3.76RR667 pKa = 11.84DD668 pKa = 3.84AVTNEE673 pKa = 3.36IADD676 pKa = 3.29ISKK679 pKa = 10.24YY680 pKa = 10.66RR681 pKa = 11.84NTNFPGLASNTIYY694 pKa = 10.55IKK696 pKa = 10.43IINKK700 pKa = 9.41INNNCTGLSKK710 pKa = 10.92LFVRR714 pKa = 11.84VLPLPSFDD722 pKa = 3.43VTSPQILCLNNLSSIEE738 pKa = 4.04AEE740 pKa = 4.5SPGDD744 pKa = 3.37TYY746 pKa = 11.42AYY748 pKa = 9.07EE749 pKa = 3.88WTRR752 pKa = 11.84NGNATVIGNNQTLNLTQGGEE772 pKa = 4.34YY773 pKa = 10.26KK774 pKa = 9.46VTAINTTTLCKK785 pKa = 9.62RR786 pKa = 11.84SRR788 pKa = 11.84IIFVNEE794 pKa = 3.93SIIATINQNDD804 pKa = 3.51VSIIDD809 pKa = 4.64DD810 pKa = 3.88SDD812 pKa = 4.04NNTITINNDD821 pKa = 2.84TGNLGIGAYY830 pKa = 9.42EE831 pKa = 4.52FALQDD836 pKa = 3.33EE837 pKa = 4.92NGQIVQDD844 pKa = 4.09YY845 pKa = 9.8QDD847 pKa = 3.45EE848 pKa = 4.58PKK850 pKa = 10.66FEE852 pKa = 4.07NLQGGIYY859 pKa = 9.58TILVRR864 pKa = 11.84DD865 pKa = 4.09KK866 pKa = 10.02NNCGIAEE873 pKa = 4.48LDD875 pKa = 3.65VSVLEE880 pKa = 4.7FPDD883 pKa = 4.53FFTPNEE889 pKa = 3.96DD890 pKa = 4.96SINDD894 pKa = 3.36TWNVKK899 pKa = 8.94GANSLFYY906 pKa = 9.56PSNSIYY912 pKa = 10.61IFDD915 pKa = 4.41RR916 pKa = 11.84YY917 pKa = 10.64GKK919 pKa = 10.32LLTSLKK925 pKa = 10.46VDD927 pKa = 3.49DD928 pKa = 4.45QGWNGTYY935 pKa = 10.36NGNALPSNDD944 pKa = 2.8YY945 pKa = 9.62WFNIEE950 pKa = 3.88LTDD953 pKa = 4.13RR954 pKa = 11.84NGNVITRR961 pKa = 11.84KK962 pKa = 9.6GHH964 pKa = 6.3FSLLLDD970 pKa = 3.79
Molecular weight: 105.75 kDa
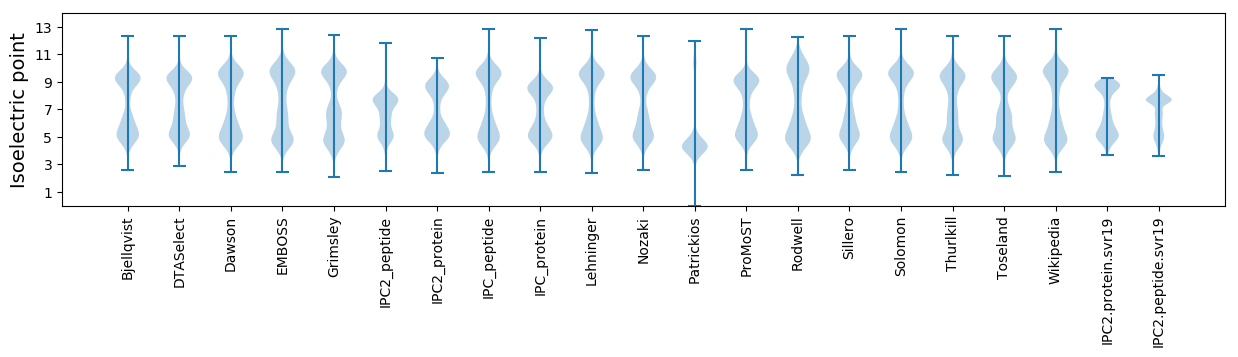
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R1A964|A0A4R1A964_9FLAO DUF1508 domain-containing protein OS=Tenacibaculum sp. M341 OX=2530339 GN=EYW44_09960 PE=4 SV=1
EEE2 pKa = 4.63FNAKKK7 pKa = 9.26KKK9 pKa = 10.71FRR11 pKa = 11.84AQFRR15 pKa = 11.84GVRR18 pKa = 11.84NVEEE22 pKa = 3.75FLYYY26 pKa = 10.6LTQIFA
EEE2 pKa = 4.63FNAKKK7 pKa = 9.26KKK9 pKa = 10.71FRR11 pKa = 11.84AQFRR15 pKa = 11.84GVRR18 pKa = 11.84NVEEE22 pKa = 3.75FLYYY26 pKa = 10.6LTQIFA
Molecular weight: 3.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1484396 |
22 |
8406 |
349.0 |
39.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.595 ± 0.036 | 0.754 ± 0.014 |
5.538 ± 0.031 | 6.727 ± 0.042 |
5.372 ± 0.031 | 6.067 ± 0.039 |
1.66 ± 0.018 | 8.171 ± 0.041 |
8.504 ± 0.064 | 9.025 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.863 ± 0.018 | 6.989 ± 0.05 |
3.096 ± 0.023 | 3.192 ± 0.022 |
3.198 ± 0.024 | 6.823 ± 0.038 |
6.049 ± 0.051 | 6.179 ± 0.032 |
1.035 ± 0.012 | 4.164 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
