
Mink coronavirus strain WD1133
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Minacovirus; unclassified Minacovirus
Average proteome isoelectric point is 7.45
Get precalculated fractions of proteins
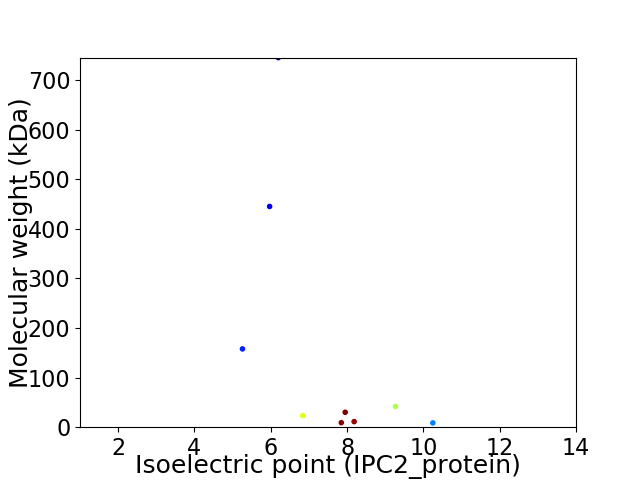
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
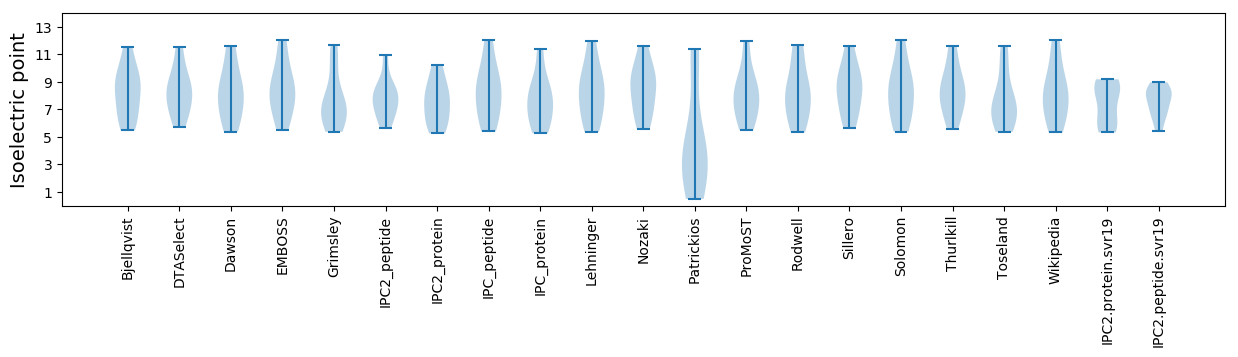
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D9J205|D9J205_9ALPC Envelope small membrane protein OS=Mink coronavirus strain WD1133 OX=766792 GN=E PE=3 SV=1
MM1 pKa = 7.87RR2 pKa = 11.84KK3 pKa = 8.29MLLILVLIPCVLGDD17 pKa = 4.58LFPCRR22 pKa = 11.84TSINSTIGNHH32 pKa = 5.97WYY34 pKa = 10.72LIDD37 pKa = 4.27NFVKK41 pKa = 10.46NYY43 pKa = 10.33SVSLPSNSDD52 pKa = 3.34VVLGDD57 pKa = 3.26YY58 pKa = 10.61FPTVEE63 pKa = 4.18PWHH66 pKa = 6.28NCLLEE71 pKa = 4.16QTNSTVIFDD80 pKa = 4.37GLRR83 pKa = 11.84ALYY86 pKa = 9.33WDD88 pKa = 4.1YY89 pKa = 11.93NNGQVPVRR97 pKa = 11.84RR98 pKa = 11.84YY99 pKa = 10.02LAVAVYY105 pKa = 10.0GDD107 pKa = 4.25PYY109 pKa = 10.74SVSVYY114 pKa = 8.42FTLDD118 pKa = 3.25YY119 pKa = 10.8EE120 pKa = 5.05GPRR123 pKa = 11.84GAKK126 pKa = 9.11ICLCKK131 pKa = 10.58GMPQPTHH138 pKa = 6.4ISGEE142 pKa = 4.21ANCWSSQCKK151 pKa = 9.37LNHH154 pKa = 6.13YY155 pKa = 10.06FPICNNTDD163 pKa = 2.95HH164 pKa = 7.17CGHH167 pKa = 6.94FLYY170 pKa = 10.64GLKK173 pKa = 9.89WSQTEE178 pKa = 4.02LFAYY182 pKa = 10.43LYY184 pKa = 10.73GYY186 pKa = 9.96VYY188 pKa = 10.61NIKK191 pKa = 10.36LVNHH195 pKa = 6.93WFNNASLALMRR206 pKa = 11.84AASVGQEE213 pKa = 3.99GWWFNPVRR221 pKa = 11.84NLTYY225 pKa = 10.24YY226 pKa = 10.4SVYY229 pKa = 10.3KK230 pKa = 10.43NSSSTIVQNCTGNCEE245 pKa = 3.95EE246 pKa = 4.31YY247 pKa = 11.19ANNIFSTEE255 pKa = 3.76PGGIIPEE262 pKa = 4.11GFSFNNWFVLTNDD275 pKa = 3.19STVLDD280 pKa = 3.84GRR282 pKa = 11.84FLTRR286 pKa = 11.84QPLLVNCLWPVPSFSEE302 pKa = 4.18ISQEE306 pKa = 4.03FCFASADD313 pKa = 3.6FTQCNGYY320 pKa = 9.69KK321 pKa = 10.34LNSTADD327 pKa = 3.52VIRR330 pKa = 11.84FNLNFTHH337 pKa = 5.99EE338 pKa = 4.17TAAASGTTFFEE349 pKa = 5.07LNTTGGVVLYY359 pKa = 9.41ISCYY363 pKa = 7.7NQSRR367 pKa = 11.84TEE369 pKa = 4.0AQVSEE374 pKa = 4.37GFVPFGNHH382 pKa = 5.91EE383 pKa = 4.26GSLYY387 pKa = 10.72CYY389 pKa = 10.34VSYY392 pKa = 11.57NEE394 pKa = 4.17TLQKK398 pKa = 10.44FLGVLPPSVKK408 pKa = 10.27EE409 pKa = 3.62IAISKK414 pKa = 10.69DD415 pKa = 2.58GGFYY419 pKa = 10.48INGYY423 pKa = 10.47NYY425 pKa = 10.1FQTFPIDD432 pKa = 4.1CISFNLTTGNTGAFWTIAYY451 pKa = 7.72TSYY454 pKa = 10.56TDD456 pKa = 3.61VMVDD460 pKa = 3.61VEE462 pKa = 4.15NTAIKK467 pKa = 10.2RR468 pKa = 11.84VIYY471 pKa = 10.15CNSHH475 pKa = 6.8INDD478 pKa = 3.1IRR480 pKa = 11.84CNQLTPSLPDD490 pKa = 2.79GFYY493 pKa = 10.32PVSPKK498 pKa = 10.76AIGFINKK505 pKa = 8.24TFVTLPANFDD515 pKa = 3.43HH516 pKa = 7.08VYY518 pKa = 11.08VNITGNVNLKK528 pKa = 10.46AEE530 pKa = 4.3RR531 pKa = 11.84GRR533 pKa = 11.84PYY535 pKa = 11.28NNGGNATLSFQGCIAVSQFTVNLNHH560 pKa = 6.89TCTITNAGSSGCTNAYY576 pKa = 7.31EE577 pKa = 4.29ANVYY581 pKa = 7.1MTSGSCPFAFDD592 pKa = 3.76RR593 pKa = 11.84LNNHH597 pKa = 5.58MSFSKK602 pKa = 10.76LCFSTVPLGDD612 pKa = 3.83DD613 pKa = 3.64CRR615 pKa = 11.84LDD617 pKa = 3.37LTVQTRR623 pKa = 11.84YY624 pKa = 7.08FTGVFAHH631 pKa = 6.54VYY633 pKa = 9.21VSYY636 pKa = 11.19KK637 pKa = 10.32FGLDD641 pKa = 2.87IVGLPTSDD649 pKa = 5.14AGLKK653 pKa = 10.16DD654 pKa = 4.12LSVLHH659 pKa = 6.59LNVCTEE665 pKa = 3.88YY666 pKa = 11.03NVYY669 pKa = 10.29GFSGTGIIRR678 pKa = 11.84QTNQSIPGGLYY689 pKa = 8.24YY690 pKa = 10.59TSLSGDD696 pKa = 3.35LLGFKK701 pKa = 10.35NVTTGTVYY709 pKa = 10.73SITPCEE715 pKa = 4.22VSAQAAVIDD724 pKa = 4.3GSIVGAITSVNSEE737 pKa = 4.03LLGLKK742 pKa = 9.63NHH744 pKa = 6.14ITTPYY749 pKa = 7.97FYY751 pKa = 10.62YY752 pKa = 10.66YY753 pKa = 10.65SVYY756 pKa = 10.8NYY758 pKa = 9.01TADD761 pKa = 3.3NVTRR765 pKa = 11.84SMQKK769 pKa = 10.27YY770 pKa = 9.57DD771 pKa = 3.67VNCTPVISYY780 pKa = 11.43SNMGVCANGALVFINVTHH798 pKa = 6.73TNGDD802 pKa = 3.57VQPISTGNVTIPSNFTISVQMEE824 pKa = 4.04YY825 pKa = 10.78VQVSTEE831 pKa = 4.19LVSIDD836 pKa = 3.32CARR839 pKa = 11.84YY840 pKa = 8.63VCNGNARR847 pKa = 11.84CGKK850 pKa = 9.88LLSQYY855 pKa = 10.68VSACHH860 pKa = 6.31TIEE863 pKa = 3.73QALAMGSRR871 pKa = 11.84LEE873 pKa = 4.13SMEE876 pKa = 4.5LEE878 pKa = 4.22SMISISANALKK889 pKa = 10.29LASVEE894 pKa = 4.2EE895 pKa = 4.54FNSSAMLNPVYY906 pKa = 10.55NEE908 pKa = 3.91SGNTIGGIYY917 pKa = 10.18LDD919 pKa = 4.0GLKK922 pKa = 10.68DD923 pKa = 3.19ILPRR927 pKa = 11.84KK928 pKa = 8.52NKK930 pKa = 9.76HH931 pKa = 4.91GSSRR935 pKa = 11.84STIEE939 pKa = 4.47DD940 pKa = 3.63LLFNKK945 pKa = 9.4VVTTGLGTVDD955 pKa = 4.12EE956 pKa = 4.86DD957 pKa = 4.55YY958 pKa = 11.25KK959 pKa = 11.14RR960 pKa = 11.84CTKK963 pKa = 10.68GSDD966 pKa = 3.08IADD969 pKa = 4.35LACAQYY975 pKa = 11.34YY976 pKa = 9.2NGIMVLPGVANDD988 pKa = 4.86GKK990 pKa = 10.3MSMYY994 pKa = 9.3TASLSGGISLGALGGGAVAIPFSLAVQARR1023 pKa = 11.84LNYY1026 pKa = 9.72VALQTDD1032 pKa = 3.9VLQRR1036 pKa = 11.84NQEE1039 pKa = 3.65ILAASFNQAIGNITIALGKK1058 pKa = 9.82VNNAIYY1064 pKa = 8.4QTSQSLSTVAQALTKK1079 pKa = 10.33VQDD1082 pKa = 3.72VVNSQGKK1089 pKa = 9.06ALNHH1093 pKa = 6.2LTLQLQNNFQAISSSIQDD1111 pKa = 3.17IYY1113 pKa = 11.73YY1114 pKa = 10.85KK1115 pKa = 10.67LDD1117 pKa = 5.03DD1118 pKa = 4.46INADD1122 pKa = 3.5AQVDD1126 pKa = 3.81RR1127 pKa = 11.84LITGRR1132 pKa = 11.84LAALNAFVTQTLTRR1146 pKa = 11.84QAEE1149 pKa = 4.39VRR1151 pKa = 11.84ASRR1154 pKa = 11.84QLAKK1158 pKa = 10.58EE1159 pKa = 3.99KK1160 pKa = 10.98VNEE1163 pKa = 4.29CVRR1166 pKa = 11.84SQSSRR1171 pKa = 11.84FGFCGNGTHH1180 pKa = 7.6LFSLTNAAPNGMVFFHH1196 pKa = 6.23TVLVPTAYY1204 pKa = 8.26EE1205 pKa = 4.24TVTAWSGICASDD1217 pKa = 3.87DD1218 pKa = 3.36SRR1220 pKa = 11.84TFGLIVKK1227 pKa = 9.68DD1228 pKa = 3.7VSLTLFRR1235 pKa = 11.84NHH1237 pKa = 7.22DD1238 pKa = 3.7GKK1240 pKa = 11.2FYY1242 pKa = 9.39LTPRR1246 pKa = 11.84TMYY1249 pKa = 9.62QPRR1252 pKa = 11.84VATSADD1258 pKa = 3.12FVQIVDD1264 pKa = 3.89CDD1266 pKa = 3.88VLFVNATILEE1276 pKa = 4.42LPSIVPDD1283 pKa = 3.83YY1284 pKa = 11.26IDD1286 pKa = 3.69INKK1289 pKa = 8.34TVQDD1293 pKa = 4.57LLDD1296 pKa = 3.85SYY1298 pKa = 10.94KK1299 pKa = 10.87PNWTVPEE1306 pKa = 4.33FSLDD1310 pKa = 3.4IFNQTYY1316 pKa = 11.1LNITNEE1322 pKa = 3.9INDD1325 pKa = 3.86LEE1327 pKa = 4.24NRR1329 pKa = 11.84SVVLYY1334 pKa = 8.17NTTKK1338 pKa = 10.47EE1339 pKa = 4.08LEE1341 pKa = 4.19LLIQSINNTLVDD1353 pKa = 4.7LEE1355 pKa = 4.39WLNKK1359 pKa = 9.78IEE1361 pKa = 5.23TYY1363 pKa = 10.77VKK1365 pKa = 8.7WPWYY1369 pKa = 8.59VWVLIALIFLFVLPMLLFCCLSTGCCGCCGCLSSCAAGCCKK1410 pKa = 10.4YY1411 pKa = 10.81GCSRR1415 pKa = 11.84DD1416 pKa = 3.43LSRR1419 pKa = 11.84YY1420 pKa = 8.12EE1421 pKa = 4.39PIEE1424 pKa = 4.11KK1425 pKa = 10.11VHH1427 pKa = 6.2VNN1429 pKa = 3.28
MM1 pKa = 7.87RR2 pKa = 11.84KK3 pKa = 8.29MLLILVLIPCVLGDD17 pKa = 4.58LFPCRR22 pKa = 11.84TSINSTIGNHH32 pKa = 5.97WYY34 pKa = 10.72LIDD37 pKa = 4.27NFVKK41 pKa = 10.46NYY43 pKa = 10.33SVSLPSNSDD52 pKa = 3.34VVLGDD57 pKa = 3.26YY58 pKa = 10.61FPTVEE63 pKa = 4.18PWHH66 pKa = 6.28NCLLEE71 pKa = 4.16QTNSTVIFDD80 pKa = 4.37GLRR83 pKa = 11.84ALYY86 pKa = 9.33WDD88 pKa = 4.1YY89 pKa = 11.93NNGQVPVRR97 pKa = 11.84RR98 pKa = 11.84YY99 pKa = 10.02LAVAVYY105 pKa = 10.0GDD107 pKa = 4.25PYY109 pKa = 10.74SVSVYY114 pKa = 8.42FTLDD118 pKa = 3.25YY119 pKa = 10.8EE120 pKa = 5.05GPRR123 pKa = 11.84GAKK126 pKa = 9.11ICLCKK131 pKa = 10.58GMPQPTHH138 pKa = 6.4ISGEE142 pKa = 4.21ANCWSSQCKK151 pKa = 9.37LNHH154 pKa = 6.13YY155 pKa = 10.06FPICNNTDD163 pKa = 2.95HH164 pKa = 7.17CGHH167 pKa = 6.94FLYY170 pKa = 10.64GLKK173 pKa = 9.89WSQTEE178 pKa = 4.02LFAYY182 pKa = 10.43LYY184 pKa = 10.73GYY186 pKa = 9.96VYY188 pKa = 10.61NIKK191 pKa = 10.36LVNHH195 pKa = 6.93WFNNASLALMRR206 pKa = 11.84AASVGQEE213 pKa = 3.99GWWFNPVRR221 pKa = 11.84NLTYY225 pKa = 10.24YY226 pKa = 10.4SVYY229 pKa = 10.3KK230 pKa = 10.43NSSSTIVQNCTGNCEE245 pKa = 3.95EE246 pKa = 4.31YY247 pKa = 11.19ANNIFSTEE255 pKa = 3.76PGGIIPEE262 pKa = 4.11GFSFNNWFVLTNDD275 pKa = 3.19STVLDD280 pKa = 3.84GRR282 pKa = 11.84FLTRR286 pKa = 11.84QPLLVNCLWPVPSFSEE302 pKa = 4.18ISQEE306 pKa = 4.03FCFASADD313 pKa = 3.6FTQCNGYY320 pKa = 9.69KK321 pKa = 10.34LNSTADD327 pKa = 3.52VIRR330 pKa = 11.84FNLNFTHH337 pKa = 5.99EE338 pKa = 4.17TAAASGTTFFEE349 pKa = 5.07LNTTGGVVLYY359 pKa = 9.41ISCYY363 pKa = 7.7NQSRR367 pKa = 11.84TEE369 pKa = 4.0AQVSEE374 pKa = 4.37GFVPFGNHH382 pKa = 5.91EE383 pKa = 4.26GSLYY387 pKa = 10.72CYY389 pKa = 10.34VSYY392 pKa = 11.57NEE394 pKa = 4.17TLQKK398 pKa = 10.44FLGVLPPSVKK408 pKa = 10.27EE409 pKa = 3.62IAISKK414 pKa = 10.69DD415 pKa = 2.58GGFYY419 pKa = 10.48INGYY423 pKa = 10.47NYY425 pKa = 10.1FQTFPIDD432 pKa = 4.1CISFNLTTGNTGAFWTIAYY451 pKa = 7.72TSYY454 pKa = 10.56TDD456 pKa = 3.61VMVDD460 pKa = 3.61VEE462 pKa = 4.15NTAIKK467 pKa = 10.2RR468 pKa = 11.84VIYY471 pKa = 10.15CNSHH475 pKa = 6.8INDD478 pKa = 3.1IRR480 pKa = 11.84CNQLTPSLPDD490 pKa = 2.79GFYY493 pKa = 10.32PVSPKK498 pKa = 10.76AIGFINKK505 pKa = 8.24TFVTLPANFDD515 pKa = 3.43HH516 pKa = 7.08VYY518 pKa = 11.08VNITGNVNLKK528 pKa = 10.46AEE530 pKa = 4.3RR531 pKa = 11.84GRR533 pKa = 11.84PYY535 pKa = 11.28NNGGNATLSFQGCIAVSQFTVNLNHH560 pKa = 6.89TCTITNAGSSGCTNAYY576 pKa = 7.31EE577 pKa = 4.29ANVYY581 pKa = 7.1MTSGSCPFAFDD592 pKa = 3.76RR593 pKa = 11.84LNNHH597 pKa = 5.58MSFSKK602 pKa = 10.76LCFSTVPLGDD612 pKa = 3.83DD613 pKa = 3.64CRR615 pKa = 11.84LDD617 pKa = 3.37LTVQTRR623 pKa = 11.84YY624 pKa = 7.08FTGVFAHH631 pKa = 6.54VYY633 pKa = 9.21VSYY636 pKa = 11.19KK637 pKa = 10.32FGLDD641 pKa = 2.87IVGLPTSDD649 pKa = 5.14AGLKK653 pKa = 10.16DD654 pKa = 4.12LSVLHH659 pKa = 6.59LNVCTEE665 pKa = 3.88YY666 pKa = 11.03NVYY669 pKa = 10.29GFSGTGIIRR678 pKa = 11.84QTNQSIPGGLYY689 pKa = 8.24YY690 pKa = 10.59TSLSGDD696 pKa = 3.35LLGFKK701 pKa = 10.35NVTTGTVYY709 pKa = 10.73SITPCEE715 pKa = 4.22VSAQAAVIDD724 pKa = 4.3GSIVGAITSVNSEE737 pKa = 4.03LLGLKK742 pKa = 9.63NHH744 pKa = 6.14ITTPYY749 pKa = 7.97FYY751 pKa = 10.62YY752 pKa = 10.66YY753 pKa = 10.65SVYY756 pKa = 10.8NYY758 pKa = 9.01TADD761 pKa = 3.3NVTRR765 pKa = 11.84SMQKK769 pKa = 10.27YY770 pKa = 9.57DD771 pKa = 3.67VNCTPVISYY780 pKa = 11.43SNMGVCANGALVFINVTHH798 pKa = 6.73TNGDD802 pKa = 3.57VQPISTGNVTIPSNFTISVQMEE824 pKa = 4.04YY825 pKa = 10.78VQVSTEE831 pKa = 4.19LVSIDD836 pKa = 3.32CARR839 pKa = 11.84YY840 pKa = 8.63VCNGNARR847 pKa = 11.84CGKK850 pKa = 9.88LLSQYY855 pKa = 10.68VSACHH860 pKa = 6.31TIEE863 pKa = 3.73QALAMGSRR871 pKa = 11.84LEE873 pKa = 4.13SMEE876 pKa = 4.5LEE878 pKa = 4.22SMISISANALKK889 pKa = 10.29LASVEE894 pKa = 4.2EE895 pKa = 4.54FNSSAMLNPVYY906 pKa = 10.55NEE908 pKa = 3.91SGNTIGGIYY917 pKa = 10.18LDD919 pKa = 4.0GLKK922 pKa = 10.68DD923 pKa = 3.19ILPRR927 pKa = 11.84KK928 pKa = 8.52NKK930 pKa = 9.76HH931 pKa = 4.91GSSRR935 pKa = 11.84STIEE939 pKa = 4.47DD940 pKa = 3.63LLFNKK945 pKa = 9.4VVTTGLGTVDD955 pKa = 4.12EE956 pKa = 4.86DD957 pKa = 4.55YY958 pKa = 11.25KK959 pKa = 11.14RR960 pKa = 11.84CTKK963 pKa = 10.68GSDD966 pKa = 3.08IADD969 pKa = 4.35LACAQYY975 pKa = 11.34YY976 pKa = 9.2NGIMVLPGVANDD988 pKa = 4.86GKK990 pKa = 10.3MSMYY994 pKa = 9.3TASLSGGISLGALGGGAVAIPFSLAVQARR1023 pKa = 11.84LNYY1026 pKa = 9.72VALQTDD1032 pKa = 3.9VLQRR1036 pKa = 11.84NQEE1039 pKa = 3.65ILAASFNQAIGNITIALGKK1058 pKa = 9.82VNNAIYY1064 pKa = 8.4QTSQSLSTVAQALTKK1079 pKa = 10.33VQDD1082 pKa = 3.72VVNSQGKK1089 pKa = 9.06ALNHH1093 pKa = 6.2LTLQLQNNFQAISSSIQDD1111 pKa = 3.17IYY1113 pKa = 11.73YY1114 pKa = 10.85KK1115 pKa = 10.67LDD1117 pKa = 5.03DD1118 pKa = 4.46INADD1122 pKa = 3.5AQVDD1126 pKa = 3.81RR1127 pKa = 11.84LITGRR1132 pKa = 11.84LAALNAFVTQTLTRR1146 pKa = 11.84QAEE1149 pKa = 4.39VRR1151 pKa = 11.84ASRR1154 pKa = 11.84QLAKK1158 pKa = 10.58EE1159 pKa = 3.99KK1160 pKa = 10.98VNEE1163 pKa = 4.29CVRR1166 pKa = 11.84SQSSRR1171 pKa = 11.84FGFCGNGTHH1180 pKa = 7.6LFSLTNAAPNGMVFFHH1196 pKa = 6.23TVLVPTAYY1204 pKa = 8.26EE1205 pKa = 4.24TVTAWSGICASDD1217 pKa = 3.87DD1218 pKa = 3.36SRR1220 pKa = 11.84TFGLIVKK1227 pKa = 9.68DD1228 pKa = 3.7VSLTLFRR1235 pKa = 11.84NHH1237 pKa = 7.22DD1238 pKa = 3.7GKK1240 pKa = 11.2FYY1242 pKa = 9.39LTPRR1246 pKa = 11.84TMYY1249 pKa = 9.62QPRR1252 pKa = 11.84VATSADD1258 pKa = 3.12FVQIVDD1264 pKa = 3.89CDD1266 pKa = 3.88VLFVNATILEE1276 pKa = 4.42LPSIVPDD1283 pKa = 3.83YY1284 pKa = 11.26IDD1286 pKa = 3.69INKK1289 pKa = 8.34TVQDD1293 pKa = 4.57LLDD1296 pKa = 3.85SYY1298 pKa = 10.94KK1299 pKa = 10.87PNWTVPEE1306 pKa = 4.33FSLDD1310 pKa = 3.4IFNQTYY1316 pKa = 11.1LNITNEE1322 pKa = 3.9INDD1325 pKa = 3.86LEE1327 pKa = 4.24NRR1329 pKa = 11.84SVVLYY1334 pKa = 8.17NTTKK1338 pKa = 10.47EE1339 pKa = 4.08LEE1341 pKa = 4.19LLIQSINNTLVDD1353 pKa = 4.7LEE1355 pKa = 4.39WLNKK1359 pKa = 9.78IEE1361 pKa = 5.23TYY1363 pKa = 10.77VKK1365 pKa = 8.7WPWYY1369 pKa = 8.59VWVLIALIFLFVLPMLLFCCLSTGCCGCCGCLSSCAAGCCKK1410 pKa = 10.4YY1411 pKa = 10.81GCSRR1415 pKa = 11.84DD1416 pKa = 3.43LSRR1419 pKa = 11.84YY1420 pKa = 8.12EE1421 pKa = 4.39PIEE1424 pKa = 4.11KK1425 pKa = 10.11VHH1427 pKa = 6.2VNN1429 pKa = 3.28
Molecular weight: 157.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D9J210|D9J210_9ALPC Accessory protein 7b OS=Mink coronavirus strain WD1133 OX=766792 PE=4 SV=1
MM1 pKa = 7.86RR2 pKa = 11.84FNLITLIPGSVWFKK16 pKa = 10.69LPRR19 pKa = 11.84PFKK22 pKa = 8.89NWIVAKK28 pKa = 9.87VKK30 pKa = 10.58RR31 pKa = 11.84KK32 pKa = 10.15VPTCRR37 pKa = 11.84SIKK40 pKa = 8.29TDD42 pKa = 3.06YY43 pKa = 10.32RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84AIYY49 pKa = 10.08NLRR52 pKa = 11.84IKK54 pKa = 10.42NASTNSCNSFSSSSCFRR71 pKa = 11.84NKK73 pKa = 10.29
MM1 pKa = 7.86RR2 pKa = 11.84FNLITLIPGSVWFKK16 pKa = 10.69LPRR19 pKa = 11.84PFKK22 pKa = 8.89NWIVAKK28 pKa = 9.87VKK30 pKa = 10.58RR31 pKa = 11.84KK32 pKa = 10.15VPTCRR37 pKa = 11.84SIKK40 pKa = 8.29TDD42 pKa = 3.06YY43 pKa = 10.32RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84AIYY49 pKa = 10.08NLRR52 pKa = 11.84IKK54 pKa = 10.42NASTNSCNSFSSSSCFRR71 pKa = 11.84NKK73 pKa = 10.29
Molecular weight: 8.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13206 |
73 |
6670 |
1467.3 |
163.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.247 ± 0.14 | 3.589 ± 0.276 |
5.498 ± 0.514 | 3.953 ± 0.192 |
5.225 ± 0.185 | 6.558 ± 0.182 |
1.833 ± 0.118 | 5.369 ± 0.295 |
6.05 ± 0.669 | 8.595 ± 0.547 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.105 ± 0.199 | 6.179 ± 0.596 |
3.355 ± 0.206 | 2.787 ± 0.291 |
3.249 ± 0.412 | 7.065 ± 0.414 |
6.171 ± 0.334 | 9.935 ± 0.612 |
1.227 ± 0.22 | 5.013 ± 0.254 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
