
Bovine faeces associated smacovirus 6
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Bostasmacovirus; Bostasmacovirus bovas1
Average proteome isoelectric point is 5.43
Get precalculated fractions of proteins
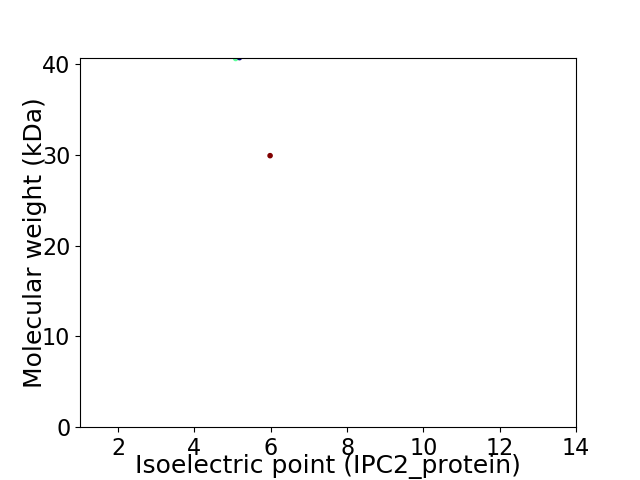
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A168MFS2|A0A168MFS2_9VIRU Putative capsid protein OS=Bovine faeces associated smacovirus 6 OX=1843754 PE=4 SV=1
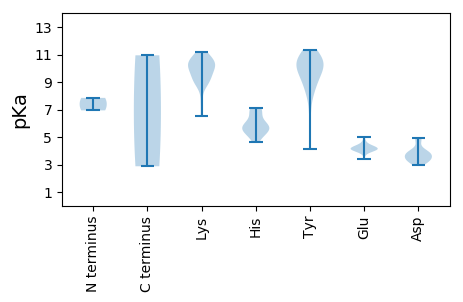
MM1 pKa = 6.94ATQYY5 pKa = 11.34AKK7 pKa = 10.44SSYY10 pKa = 10.34SEE12 pKa = 4.34VYY14 pKa = 10.84DD15 pKa = 3.24MHH17 pKa = 7.14TEE19 pKa = 4.32FGKK22 pKa = 7.6TTLIGIHH29 pKa = 6.07TPTTNRR35 pKa = 11.84WQKK38 pKa = 9.8YY39 pKa = 6.22MRR41 pKa = 11.84GFCHH45 pKa = 5.38QFKK48 pKa = 10.58KK49 pKa = 10.64FRR51 pKa = 11.84YY52 pKa = 8.44QGASLSFVPVATLPVDD68 pKa = 3.81PLAVSYY74 pKa = 10.74EE75 pKa = 4.07GGEE78 pKa = 4.04AQIDD82 pKa = 3.82PRR84 pKa = 11.84DD85 pKa = 3.73MVNPIIHH92 pKa = 6.59CGMRR96 pKa = 11.84GEE98 pKa = 4.77SMGKK102 pKa = 8.68YY103 pKa = 9.53LDD105 pKa = 4.55EE106 pKa = 4.35IFHH109 pKa = 6.92FNLEE113 pKa = 4.25HH114 pKa = 7.0KK115 pKa = 10.56LNSVDD120 pKa = 3.31VANVVNEE127 pKa = 4.13DD128 pKa = 4.92LIADD132 pKa = 4.84LEE134 pKa = 4.26NMYY137 pKa = 8.89YY138 pKa = 9.66TALSSPAFKK147 pKa = 10.32KK148 pKa = 10.05SHH150 pKa = 5.8PQQGFRR156 pKa = 11.84KK157 pKa = 9.38SGLHH161 pKa = 5.44PMVYY165 pKa = 10.37SLATDD170 pKa = 3.68RR171 pKa = 11.84PKK173 pKa = 11.12NFSLPDD179 pKa = 3.42TDD181 pKa = 3.75TDD183 pKa = 3.91ADD185 pKa = 4.1ALPSSTAGPLSGEE198 pKa = 4.14PGGNATDD205 pKa = 3.73GFYY208 pKa = 11.18GSFGVPYY215 pKa = 9.87AQARR219 pKa = 11.84MQVDD223 pKa = 3.35YY224 pKa = 11.0GFNGVEE230 pKa = 4.29TQRR233 pKa = 11.84QFPQLFTNHH242 pKa = 5.92AQSLGWLDD250 pKa = 3.22TMQNYY255 pKa = 10.43SIYY258 pKa = 10.26SEE260 pKa = 4.38LGGVPAGTTEE270 pKa = 3.88QYY272 pKa = 10.0LTGLPKK278 pKa = 9.31IYY280 pKa = 9.16MYY282 pKa = 10.95FVMLPPAYY290 pKa = 8.7KK291 pKa = 8.93TEE293 pKa = 4.05LYY295 pKa = 9.83FRR297 pKa = 11.84VIIRR301 pKa = 11.84HH302 pKa = 4.66FFEE305 pKa = 4.4FKK307 pKa = 10.05EE308 pKa = 4.13YY309 pKa = 11.0RR310 pKa = 11.84NMIQPFAMEE319 pKa = 4.97TINGSAYY326 pKa = 10.68DD327 pKa = 4.12GVLEE331 pKa = 4.2TAPVDD336 pKa = 3.34QAMALRR342 pKa = 11.84MEE344 pKa = 4.49NATLDD349 pKa = 3.87VVNGEE354 pKa = 4.2ASLMSDD360 pKa = 3.96GSSS363 pKa = 2.87
MM1 pKa = 6.94ATQYY5 pKa = 11.34AKK7 pKa = 10.44SSYY10 pKa = 10.34SEE12 pKa = 4.34VYY14 pKa = 10.84DD15 pKa = 3.24MHH17 pKa = 7.14TEE19 pKa = 4.32FGKK22 pKa = 7.6TTLIGIHH29 pKa = 6.07TPTTNRR35 pKa = 11.84WQKK38 pKa = 9.8YY39 pKa = 6.22MRR41 pKa = 11.84GFCHH45 pKa = 5.38QFKK48 pKa = 10.58KK49 pKa = 10.64FRR51 pKa = 11.84YY52 pKa = 8.44QGASLSFVPVATLPVDD68 pKa = 3.81PLAVSYY74 pKa = 10.74EE75 pKa = 4.07GGEE78 pKa = 4.04AQIDD82 pKa = 3.82PRR84 pKa = 11.84DD85 pKa = 3.73MVNPIIHH92 pKa = 6.59CGMRR96 pKa = 11.84GEE98 pKa = 4.77SMGKK102 pKa = 8.68YY103 pKa = 9.53LDD105 pKa = 4.55EE106 pKa = 4.35IFHH109 pKa = 6.92FNLEE113 pKa = 4.25HH114 pKa = 7.0KK115 pKa = 10.56LNSVDD120 pKa = 3.31VANVVNEE127 pKa = 4.13DD128 pKa = 4.92LIADD132 pKa = 4.84LEE134 pKa = 4.26NMYY137 pKa = 8.89YY138 pKa = 9.66TALSSPAFKK147 pKa = 10.32KK148 pKa = 10.05SHH150 pKa = 5.8PQQGFRR156 pKa = 11.84KK157 pKa = 9.38SGLHH161 pKa = 5.44PMVYY165 pKa = 10.37SLATDD170 pKa = 3.68RR171 pKa = 11.84PKK173 pKa = 11.12NFSLPDD179 pKa = 3.42TDD181 pKa = 3.75TDD183 pKa = 3.91ADD185 pKa = 4.1ALPSSTAGPLSGEE198 pKa = 4.14PGGNATDD205 pKa = 3.73GFYY208 pKa = 11.18GSFGVPYY215 pKa = 9.87AQARR219 pKa = 11.84MQVDD223 pKa = 3.35YY224 pKa = 11.0GFNGVEE230 pKa = 4.29TQRR233 pKa = 11.84QFPQLFTNHH242 pKa = 5.92AQSLGWLDD250 pKa = 3.22TMQNYY255 pKa = 10.43SIYY258 pKa = 10.26SEE260 pKa = 4.38LGGVPAGTTEE270 pKa = 3.88QYY272 pKa = 10.0LTGLPKK278 pKa = 9.31IYY280 pKa = 9.16MYY282 pKa = 10.95FVMLPPAYY290 pKa = 8.7KK291 pKa = 8.93TEE293 pKa = 4.05LYY295 pKa = 9.83FRR297 pKa = 11.84VIIRR301 pKa = 11.84HH302 pKa = 4.66FFEE305 pKa = 4.4FKK307 pKa = 10.05EE308 pKa = 4.13YY309 pKa = 11.0RR310 pKa = 11.84NMIQPFAMEE319 pKa = 4.97TINGSAYY326 pKa = 10.68DD327 pKa = 4.12GVLEE331 pKa = 4.2TAPVDD336 pKa = 3.34QAMALRR342 pKa = 11.84MEE344 pKa = 4.49NATLDD349 pKa = 3.87VVNGEE354 pKa = 4.2ASLMSDD360 pKa = 3.96GSSS363 pKa = 2.87
Molecular weight: 40.6 kDa
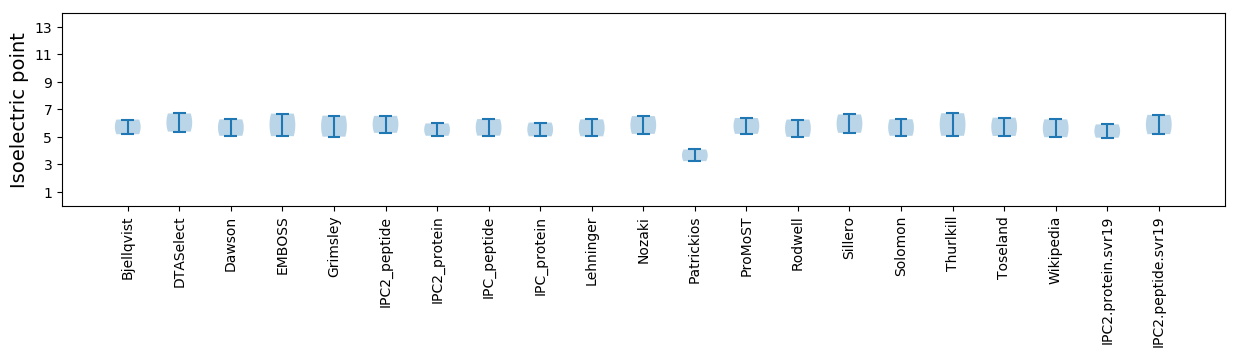
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A168MFS2|A0A168MFS2_9VIRU Putative capsid protein OS=Bovine faeces associated smacovirus 6 OX=1843754 PE=4 SV=1
MM1 pKa = 7.85SDD3 pKa = 3.61PKK5 pKa = 10.28WYY7 pKa = 10.21DD8 pKa = 2.95VTAPQAVLSAEE19 pKa = 4.03NVEE22 pKa = 4.68TILEE26 pKa = 4.01QLEE29 pKa = 4.37CEE31 pKa = 4.17RR32 pKa = 11.84YY33 pKa = 9.8AYY35 pKa = 10.34ADD37 pKa = 3.47EE38 pKa = 4.42VGEE41 pKa = 4.15GGYY44 pKa = 9.45KK45 pKa = 9.59HH46 pKa = 5.14WQIRR50 pKa = 11.84YY51 pKa = 7.43VLRR54 pKa = 11.84KK55 pKa = 9.02GSPVEE60 pKa = 3.85EE61 pKa = 4.55QIMIWSMWKK70 pKa = 9.11AHH72 pKa = 5.71VSPTHH77 pKa = 5.33VRR79 pKa = 11.84NFNYY83 pKa = 9.73IMKK86 pKa = 8.88TDD88 pKa = 3.8NYY90 pKa = 8.06FCSWEE95 pKa = 4.17SEE97 pKa = 3.99LRR99 pKa = 11.84EE100 pKa = 4.01YY101 pKa = 10.69RR102 pKa = 11.84YY103 pKa = 10.5LEE105 pKa = 3.65YY106 pKa = 10.67RR107 pKa = 11.84PWQKK111 pKa = 9.45YY112 pKa = 4.1TLEE115 pKa = 4.54MIEE118 pKa = 3.95QQNDD122 pKa = 3.07RR123 pKa = 11.84EE124 pKa = 4.28IFVIQDD130 pKa = 3.49PLGGIGKK137 pKa = 6.53TTFAKK142 pKa = 10.54HH143 pKa = 5.67LVANHH148 pKa = 5.78KK149 pKa = 10.35AIYY152 pKa = 9.71IPPLGEE158 pKa = 3.93GQEE161 pKa = 4.12YY162 pKa = 8.47MAMAMAKK169 pKa = 9.65ARR171 pKa = 11.84EE172 pKa = 4.2GRR174 pKa = 11.84RR175 pKa = 11.84EE176 pKa = 3.81TFIIDD181 pKa = 3.37VPRR184 pKa = 11.84AEE186 pKa = 4.26SLKK189 pKa = 10.48KK190 pKa = 9.95KK191 pKa = 10.32AGMWSAVEE199 pKa = 3.91QIKK202 pKa = 11.15NGFLYY207 pKa = 10.45DD208 pKa = 4.91KK209 pKa = 10.29RR210 pKa = 11.84YY211 pKa = 8.69QWSEE215 pKa = 3.4KK216 pKa = 8.98WIKK219 pKa = 10.02SPKK222 pKa = 9.72IVVLTNEE229 pKa = 4.11TEE231 pKa = 4.08LPKK234 pKa = 10.98DD235 pKa = 3.32KK236 pKa = 10.91LSNDD240 pKa = 2.94RR241 pKa = 11.84WNVYY245 pKa = 9.4QPGNFEE251 pKa = 4.16YY252 pKa = 10.94
MM1 pKa = 7.85SDD3 pKa = 3.61PKK5 pKa = 10.28WYY7 pKa = 10.21DD8 pKa = 2.95VTAPQAVLSAEE19 pKa = 4.03NVEE22 pKa = 4.68TILEE26 pKa = 4.01QLEE29 pKa = 4.37CEE31 pKa = 4.17RR32 pKa = 11.84YY33 pKa = 9.8AYY35 pKa = 10.34ADD37 pKa = 3.47EE38 pKa = 4.42VGEE41 pKa = 4.15GGYY44 pKa = 9.45KK45 pKa = 9.59HH46 pKa = 5.14WQIRR50 pKa = 11.84YY51 pKa = 7.43VLRR54 pKa = 11.84KK55 pKa = 9.02GSPVEE60 pKa = 3.85EE61 pKa = 4.55QIMIWSMWKK70 pKa = 9.11AHH72 pKa = 5.71VSPTHH77 pKa = 5.33VRR79 pKa = 11.84NFNYY83 pKa = 9.73IMKK86 pKa = 8.88TDD88 pKa = 3.8NYY90 pKa = 8.06FCSWEE95 pKa = 4.17SEE97 pKa = 3.99LRR99 pKa = 11.84EE100 pKa = 4.01YY101 pKa = 10.69RR102 pKa = 11.84YY103 pKa = 10.5LEE105 pKa = 3.65YY106 pKa = 10.67RR107 pKa = 11.84PWQKK111 pKa = 9.45YY112 pKa = 4.1TLEE115 pKa = 4.54MIEE118 pKa = 3.95QQNDD122 pKa = 3.07RR123 pKa = 11.84EE124 pKa = 4.28IFVIQDD130 pKa = 3.49PLGGIGKK137 pKa = 6.53TTFAKK142 pKa = 10.54HH143 pKa = 5.67LVANHH148 pKa = 5.78KK149 pKa = 10.35AIYY152 pKa = 9.71IPPLGEE158 pKa = 3.93GQEE161 pKa = 4.12YY162 pKa = 8.47MAMAMAKK169 pKa = 9.65ARR171 pKa = 11.84EE172 pKa = 4.2GRR174 pKa = 11.84RR175 pKa = 11.84EE176 pKa = 3.81TFIIDD181 pKa = 3.37VPRR184 pKa = 11.84AEE186 pKa = 4.26SLKK189 pKa = 10.48KK190 pKa = 9.95KK191 pKa = 10.32AGMWSAVEE199 pKa = 3.91QIKK202 pKa = 11.15NGFLYY207 pKa = 10.45DD208 pKa = 4.91KK209 pKa = 10.29RR210 pKa = 11.84YY211 pKa = 8.69QWSEE215 pKa = 3.4KK216 pKa = 8.98WIKK219 pKa = 10.02SPKK222 pKa = 9.72IVVLTNEE229 pKa = 4.11TEE231 pKa = 4.08LPKK234 pKa = 10.98DD235 pKa = 3.32KK236 pKa = 10.91LSNDD240 pKa = 2.94RR241 pKa = 11.84WNVYY245 pKa = 9.4QPGNFEE251 pKa = 4.16YY252 pKa = 10.94
Molecular weight: 29.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
615 |
252 |
363 |
307.5 |
35.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.992 ± 0.412 | 0.65 ± 0.092 |
4.878 ± 0.583 | 7.48 ± 1.819 |
4.553 ± 1.138 | 6.829 ± 1.071 |
2.439 ± 0.292 | 4.715 ± 1.047 |
5.528 ± 1.543 | 6.829 ± 0.562 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.228 ± 0.421 | 4.39 ± 0.016 |
5.528 ± 0.491 | 4.715 ± 0.03 |
4.39 ± 0.747 | 6.179 ± 0.908 |
5.528 ± 1.0 | 5.854 ± 0.063 |
1.951 ± 1.293 | 6.341 ± 0.259 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
