
Clostridium sp. CAG:451
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; environmental samples
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
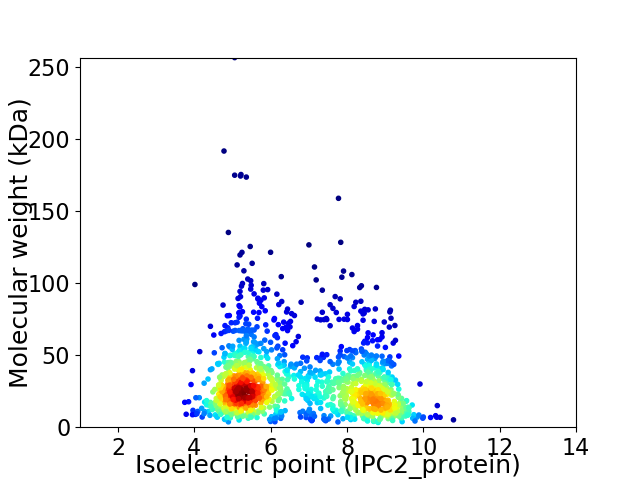
Virtual 2D-PAGE plot for 1322 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7KEG2|R7KEG2_9CLOT Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B OS=Clostridium sp. CAG:451 OX=1262809 GN=gatB PE=3 SV=1
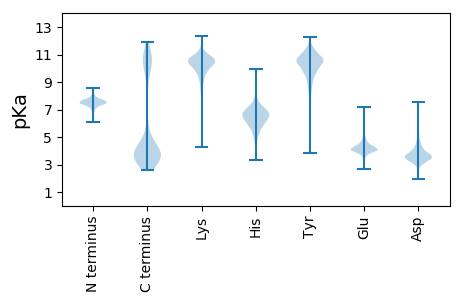
MM1 pKa = 7.71SKK3 pKa = 10.37PIYY6 pKa = 8.29ATPLSEE12 pKa = 4.33TNTTYY17 pKa = 10.85AYY19 pKa = 10.12KK20 pKa = 9.63LTIGGKK26 pKa = 10.09DD27 pKa = 3.29GIIANEE33 pKa = 3.81SMIGYY38 pKa = 9.53FYY40 pKa = 10.88EE41 pKa = 4.39FTNMTSIDD49 pKa = 4.3LSVLDD54 pKa = 3.98TSEE57 pKa = 4.07VTNMSNMFSACISLTSLDD75 pKa = 3.47VSNFDD80 pKa = 3.38TSKK83 pKa = 11.09VIDD86 pKa = 3.9MSDD89 pKa = 3.37MFDD92 pKa = 2.94QCYY95 pKa = 9.7MLTSLDD101 pKa = 3.44LRR103 pKa = 11.84NFNTSEE109 pKa = 4.21VIDD112 pKa = 3.91MSSMFSACISLTSLDD127 pKa = 3.47VSNFDD132 pKa = 3.32TSKK135 pKa = 10.6VINMSDD141 pKa = 3.26MFSEE145 pKa = 4.75CSSLTSLDD153 pKa = 3.57LKK155 pKa = 10.6EE156 pKa = 4.46SRR158 pKa = 11.84FF159 pKa = 3.79
MM1 pKa = 7.71SKK3 pKa = 10.37PIYY6 pKa = 8.29ATPLSEE12 pKa = 4.33TNTTYY17 pKa = 10.85AYY19 pKa = 10.12KK20 pKa = 9.63LTIGGKK26 pKa = 10.09DD27 pKa = 3.29GIIANEE33 pKa = 3.81SMIGYY38 pKa = 9.53FYY40 pKa = 10.88EE41 pKa = 4.39FTNMTSIDD49 pKa = 4.3LSVLDD54 pKa = 3.98TSEE57 pKa = 4.07VTNMSNMFSACISLTSLDD75 pKa = 3.47VSNFDD80 pKa = 3.38TSKK83 pKa = 11.09VIDD86 pKa = 3.9MSDD89 pKa = 3.37MFDD92 pKa = 2.94QCYY95 pKa = 9.7MLTSLDD101 pKa = 3.44LRR103 pKa = 11.84NFNTSEE109 pKa = 4.21VIDD112 pKa = 3.91MSSMFSACISLTSLDD127 pKa = 3.47VSNFDD132 pKa = 3.32TSKK135 pKa = 10.6VINMSDD141 pKa = 3.26MFSEE145 pKa = 4.75CSSLTSLDD153 pKa = 3.57LKK155 pKa = 10.6EE156 pKa = 4.46SRR158 pKa = 11.84FF159 pKa = 3.79
Molecular weight: 17.68 kDa
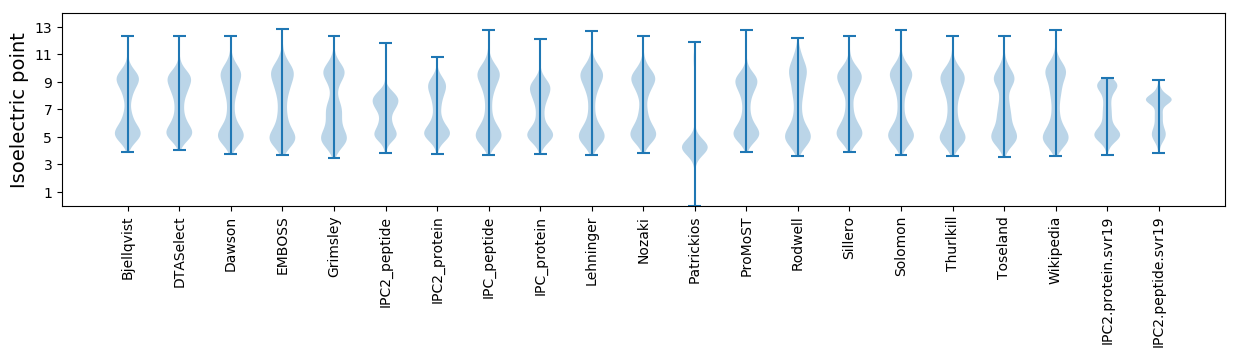
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7KB23|R7KB23_9CLOT Uncharacterized protein OS=Clostridium sp. CAG:451 OX=1262809 GN=BN663_00461 PE=4 SV=1
MM1 pKa = 7.83AIKK4 pKa = 10.31KK5 pKa = 8.03FRR7 pKa = 11.84PTTPGQRR14 pKa = 11.84NKK16 pKa = 9.66STLVNEE22 pKa = 5.47EE23 pKa = 3.96ITKK26 pKa = 8.26STPEE30 pKa = 3.66KK31 pKa = 10.8SLVVTLNKK39 pKa = 10.09KK40 pKa = 10.19AGRR43 pKa = 11.84NNQGKK48 pKa = 8.01ITVRR52 pKa = 11.84HH53 pKa = 5.14QGGGVKK59 pKa = 9.9RR60 pKa = 11.84KK61 pKa = 9.91YY62 pKa = 10.45RR63 pKa = 11.84IIDD66 pKa = 3.65FKK68 pKa = 11.18RR69 pKa = 11.84NKK71 pKa = 9.76FDD73 pKa = 3.35VEE75 pKa = 4.35GVVASIEE82 pKa = 4.01YY83 pKa = 10.59DD84 pKa = 3.46PNRR87 pKa = 11.84SANIALINYY96 pKa = 9.36LDD98 pKa = 3.67GEE100 pKa = 4.28KK101 pKa = 10.22RR102 pKa = 11.84YY103 pKa = 10.24IIAPKK108 pKa = 9.58GLKK111 pKa = 10.45VGDD114 pKa = 4.34KK115 pKa = 10.39IVAGSNADD123 pKa = 3.12IKK125 pKa = 10.11TGNALPIMNIPVGTVIHH142 pKa = 6.49NIEE145 pKa = 4.22LRR147 pKa = 11.84PGKK150 pKa = 10.2GGSLARR156 pKa = 11.84SAGSSAQILGRR167 pKa = 11.84EE168 pKa = 4.35GNYY171 pKa = 10.43VMIRR175 pKa = 11.84LSSGEE180 pKa = 3.73QRR182 pKa = 11.84RR183 pKa = 11.84VLGTCMATIGVVGNEE198 pKa = 4.07DD199 pKa = 3.66SNLVKK204 pKa = 10.51LGKK207 pKa = 10.1AGRR210 pKa = 11.84KK211 pKa = 6.0RR212 pKa = 11.84HH213 pKa = 5.13MGIRR217 pKa = 11.84PTVRR221 pKa = 11.84GSVMNPNDD229 pKa = 3.69HH230 pKa = 6.5PHH232 pKa = 6.52GGGEE236 pKa = 3.83GRR238 pKa = 11.84APIGRR243 pKa = 11.84KK244 pKa = 10.0GPVTPWGKK252 pKa = 9.06PALGYY257 pKa = 7.87KK258 pKa = 8.51TRR260 pKa = 11.84NNKK263 pKa = 8.52RR264 pKa = 11.84TDD266 pKa = 2.87KK267 pKa = 10.95FIVRR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84GSKK276 pKa = 9.95
MM1 pKa = 7.83AIKK4 pKa = 10.31KK5 pKa = 8.03FRR7 pKa = 11.84PTTPGQRR14 pKa = 11.84NKK16 pKa = 9.66STLVNEE22 pKa = 5.47EE23 pKa = 3.96ITKK26 pKa = 8.26STPEE30 pKa = 3.66KK31 pKa = 10.8SLVVTLNKK39 pKa = 10.09KK40 pKa = 10.19AGRR43 pKa = 11.84NNQGKK48 pKa = 8.01ITVRR52 pKa = 11.84HH53 pKa = 5.14QGGGVKK59 pKa = 9.9RR60 pKa = 11.84KK61 pKa = 9.91YY62 pKa = 10.45RR63 pKa = 11.84IIDD66 pKa = 3.65FKK68 pKa = 11.18RR69 pKa = 11.84NKK71 pKa = 9.76FDD73 pKa = 3.35VEE75 pKa = 4.35GVVASIEE82 pKa = 4.01YY83 pKa = 10.59DD84 pKa = 3.46PNRR87 pKa = 11.84SANIALINYY96 pKa = 9.36LDD98 pKa = 3.67GEE100 pKa = 4.28KK101 pKa = 10.22RR102 pKa = 11.84YY103 pKa = 10.24IIAPKK108 pKa = 9.58GLKK111 pKa = 10.45VGDD114 pKa = 4.34KK115 pKa = 10.39IVAGSNADD123 pKa = 3.12IKK125 pKa = 10.11TGNALPIMNIPVGTVIHH142 pKa = 6.49NIEE145 pKa = 4.22LRR147 pKa = 11.84PGKK150 pKa = 10.2GGSLARR156 pKa = 11.84SAGSSAQILGRR167 pKa = 11.84EE168 pKa = 4.35GNYY171 pKa = 10.43VMIRR175 pKa = 11.84LSSGEE180 pKa = 3.73QRR182 pKa = 11.84RR183 pKa = 11.84VLGTCMATIGVVGNEE198 pKa = 4.07DD199 pKa = 3.66SNLVKK204 pKa = 10.51LGKK207 pKa = 10.1AGRR210 pKa = 11.84KK211 pKa = 6.0RR212 pKa = 11.84HH213 pKa = 5.13MGIRR217 pKa = 11.84PTVRR221 pKa = 11.84GSVMNPNDD229 pKa = 3.69HH230 pKa = 6.5PHH232 pKa = 6.52GGGEE236 pKa = 3.83GRR238 pKa = 11.84APIGRR243 pKa = 11.84KK244 pKa = 10.0GPVTPWGKK252 pKa = 9.06PALGYY257 pKa = 7.87KK258 pKa = 8.51TRR260 pKa = 11.84NNKK263 pKa = 8.52RR264 pKa = 11.84TDD266 pKa = 2.87KK267 pKa = 10.95FIVRR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84GSKK276 pKa = 9.95
Molecular weight: 29.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
387124 |
29 |
2365 |
292.8 |
33.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.463 ± 0.057 | 1.021 ± 0.024 |
6.254 ± 0.052 | 7.207 ± 0.081 |
4.308 ± 0.054 | 5.218 ± 0.069 |
1.335 ± 0.026 | 9.131 ± 0.069 |
9.961 ± 0.082 | 9.659 ± 0.083 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.413 ± 0.031 | 6.838 ± 0.072 |
2.449 ± 0.035 | 2.214 ± 0.037 |
3.29 ± 0.047 | 6.627 ± 0.066 |
5.836 ± 0.077 | 6.092 ± 0.052 |
0.528 ± 0.015 | 5.157 ± 0.057 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
