
Streptomyces sp. 3214.6
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
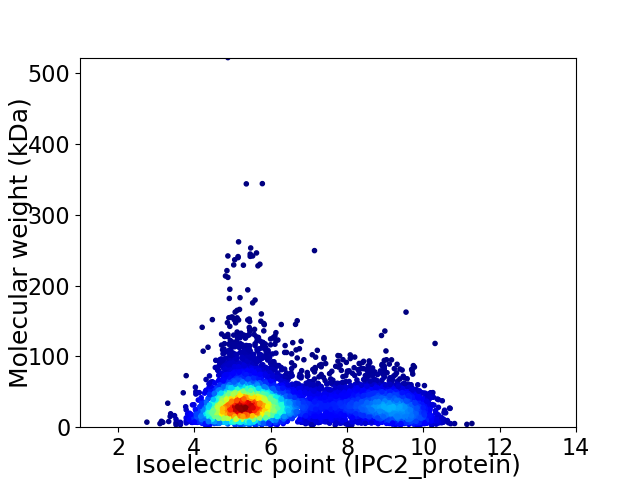
Virtual 2D-PAGE plot for 8041 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M5ZHR3|A0A1M5ZHR3_9ACTN SnoaL-like domain-containing protein OS=Streptomyces sp. 3214.6 OX=1882757 GN=SAMN05444521_5954 PE=4 SV=1
MM1 pKa = 7.54SGACGIGPSTSAYY14 pKa = 10.46AAGCTGGVDD23 pKa = 3.85SDD25 pKa = 4.32FNGDD29 pKa = 3.7GVRR32 pKa = 11.84DD33 pKa = 3.61TAIADD38 pKa = 3.63PQATVSGQEE47 pKa = 3.58AAGAIQIVLGGGKK60 pKa = 10.19GVTTLTQDD68 pKa = 3.42TPNVSDD74 pKa = 3.55VAEE77 pKa = 4.88AGDD80 pKa = 3.56QFGFSLAVYY89 pKa = 9.43DD90 pKa = 5.24ANLDD94 pKa = 3.7GCSDD98 pKa = 3.52IAVGMPYY105 pKa = 10.48EE106 pKa = 4.63DD107 pKa = 3.62VGSVQDD113 pKa = 3.74AGLVQLIYY121 pKa = 10.92GSTAGLGQGTTGSLGFRR138 pKa = 11.84QGADD142 pKa = 2.81GNMADD147 pKa = 4.1VYY149 pKa = 10.69EE150 pKa = 5.29AEE152 pKa = 4.28DD153 pKa = 3.43WVGYY157 pKa = 9.75SVAGGVSATGVPFLVIGVPGEE178 pKa = 4.26DD179 pKa = 3.38SSGLTDD185 pKa = 3.72MGLAAYY191 pKa = 9.5VAGVTPAVTSFHH203 pKa = 6.33QNSPDD208 pKa = 3.27VWHH211 pKa = 7.44DD212 pKa = 3.21AAAGNRR218 pKa = 11.84FGWAVAATGRR228 pKa = 11.84HH229 pKa = 4.78FVVGAPGAAIDD240 pKa = 4.63AADD243 pKa = 3.4QAGAVVAFKK252 pKa = 10.71PSLNADD258 pKa = 4.61NIPQPLFGIGQDD270 pKa = 3.32ATGDD274 pKa = 3.64KK275 pKa = 11.0DD276 pKa = 3.59PTAEE280 pKa = 4.11TGDD283 pKa = 3.61GFGISVAIAPYY294 pKa = 9.76RR295 pKa = 11.84PSGATATTDD304 pKa = 3.13SLVAVGVPNEE314 pKa = 4.18DD315 pKa = 3.03VGTKK319 pKa = 9.13IDD321 pKa = 3.45AGGVQLLQIAEE332 pKa = 4.35AGTVTEE338 pKa = 4.68TQWITQDD345 pKa = 3.27TADD348 pKa = 3.41VDD350 pKa = 4.1EE351 pKa = 4.63TAEE354 pKa = 3.89EE355 pKa = 3.97SDD357 pKa = 3.76YY358 pKa = 11.3FGQRR362 pKa = 11.84VAAANTNQGVVGTATTMRR380 pKa = 11.84VAVGVPGEE388 pKa = 4.12EE389 pKa = 3.7TAAAAQDD396 pKa = 3.51AGGVQIFPMLGAAGTSDD413 pKa = 3.45SWLQPGSGIPAAAAPRR429 pKa = 11.84QFAGMSVGATGSLLYY444 pKa = 10.72VGLPYY449 pKa = 10.83GPAAGRR455 pKa = 11.84AVYY458 pKa = 9.98GYY460 pKa = 10.1AWTVASGGAPSQTWKK475 pKa = 10.0PGEE478 pKa = 4.14GGIPAGAGAFGATVRR493 pKa = 4.25
MM1 pKa = 7.54SGACGIGPSTSAYY14 pKa = 10.46AAGCTGGVDD23 pKa = 3.85SDD25 pKa = 4.32FNGDD29 pKa = 3.7GVRR32 pKa = 11.84DD33 pKa = 3.61TAIADD38 pKa = 3.63PQATVSGQEE47 pKa = 3.58AAGAIQIVLGGGKK60 pKa = 10.19GVTTLTQDD68 pKa = 3.42TPNVSDD74 pKa = 3.55VAEE77 pKa = 4.88AGDD80 pKa = 3.56QFGFSLAVYY89 pKa = 9.43DD90 pKa = 5.24ANLDD94 pKa = 3.7GCSDD98 pKa = 3.52IAVGMPYY105 pKa = 10.48EE106 pKa = 4.63DD107 pKa = 3.62VGSVQDD113 pKa = 3.74AGLVQLIYY121 pKa = 10.92GSTAGLGQGTTGSLGFRR138 pKa = 11.84QGADD142 pKa = 2.81GNMADD147 pKa = 4.1VYY149 pKa = 10.69EE150 pKa = 5.29AEE152 pKa = 4.28DD153 pKa = 3.43WVGYY157 pKa = 9.75SVAGGVSATGVPFLVIGVPGEE178 pKa = 4.26DD179 pKa = 3.38SSGLTDD185 pKa = 3.72MGLAAYY191 pKa = 9.5VAGVTPAVTSFHH203 pKa = 6.33QNSPDD208 pKa = 3.27VWHH211 pKa = 7.44DD212 pKa = 3.21AAAGNRR218 pKa = 11.84FGWAVAATGRR228 pKa = 11.84HH229 pKa = 4.78FVVGAPGAAIDD240 pKa = 4.63AADD243 pKa = 3.4QAGAVVAFKK252 pKa = 10.71PSLNADD258 pKa = 4.61NIPQPLFGIGQDD270 pKa = 3.32ATGDD274 pKa = 3.64KK275 pKa = 11.0DD276 pKa = 3.59PTAEE280 pKa = 4.11TGDD283 pKa = 3.61GFGISVAIAPYY294 pKa = 9.76RR295 pKa = 11.84PSGATATTDD304 pKa = 3.13SLVAVGVPNEE314 pKa = 4.18DD315 pKa = 3.03VGTKK319 pKa = 9.13IDD321 pKa = 3.45AGGVQLLQIAEE332 pKa = 4.35AGTVTEE338 pKa = 4.68TQWITQDD345 pKa = 3.27TADD348 pKa = 3.41VDD350 pKa = 4.1EE351 pKa = 4.63TAEE354 pKa = 3.89EE355 pKa = 3.97SDD357 pKa = 3.76YY358 pKa = 11.3FGQRR362 pKa = 11.84VAAANTNQGVVGTATTMRR380 pKa = 11.84VAVGVPGEE388 pKa = 4.12EE389 pKa = 3.7TAAAAQDD396 pKa = 3.51AGGVQIFPMLGAAGTSDD413 pKa = 3.45SWLQPGSGIPAAAAPRR429 pKa = 11.84QFAGMSVGATGSLLYY444 pKa = 10.72VGLPYY449 pKa = 10.83GPAAGRR455 pKa = 11.84AVYY458 pKa = 9.98GYY460 pKa = 10.1AWTVASGGAPSQTWKK475 pKa = 10.0PGEE478 pKa = 4.14GGIPAGAGAFGATVRR493 pKa = 4.25
Molecular weight: 48.58 kDa
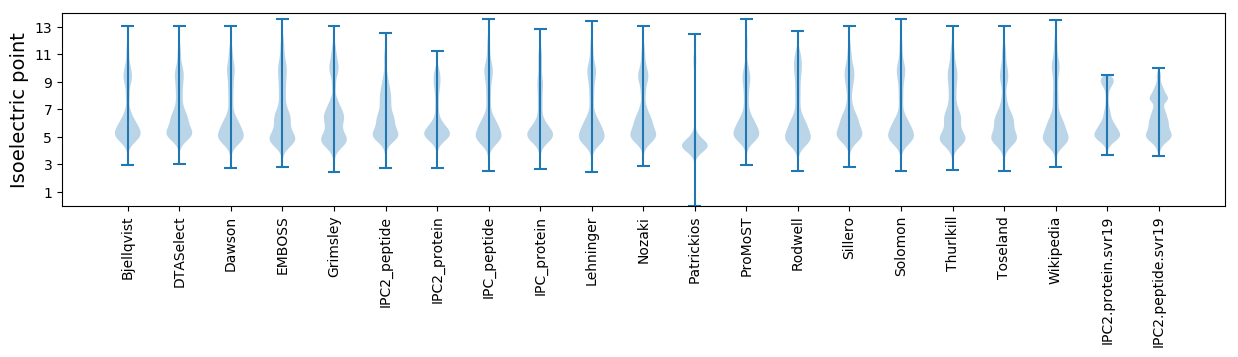
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M5YTB1|A0A1M5YTB1_9ACTN Putative peptidoglycan lipid II flippase OS=Streptomyces sp. 3214.6 OX=1882757 GN=SAMN05444521_4515 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILANRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.89GRR40 pKa = 11.84ASLSAA45 pKa = 3.83
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILANRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.89GRR40 pKa = 11.84ASLSAA45 pKa = 3.83
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2692240 |
29 |
5005 |
334.8 |
35.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.451 ± 0.04 | 0.799 ± 0.008 |
5.993 ± 0.022 | 5.567 ± 0.03 |
2.721 ± 0.016 | 9.366 ± 0.027 |
2.302 ± 0.015 | 3.144 ± 0.019 |
2.283 ± 0.025 | 10.174 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.7 ± 0.011 | 1.893 ± 0.017 |
6.081 ± 0.028 | 2.906 ± 0.017 |
7.836 ± 0.032 | 5.242 ± 0.023 |
6.382 ± 0.029 | 8.432 ± 0.026 |
1.562 ± 0.013 | 2.163 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
