
Acinetobacter phage vB_AbaM_ME3
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Metrivirus; Acinetobacter virus ME3
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
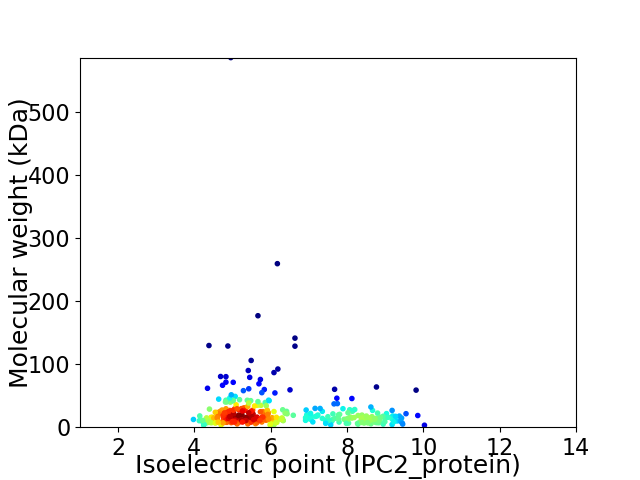
Virtual 2D-PAGE plot for 326 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A172Q054|A0A172Q054_9CAUD Putative HNH endonuclease protein OS=Acinetobacter phage vB_AbaM_ME3 OX=1837876 GN=ME3_63 PE=4 SV=1
MM1 pKa = 7.65NDD3 pKa = 3.16SEE5 pKa = 5.36VYY7 pKa = 8.19TFKK10 pKa = 10.77VRR12 pKa = 11.84YY13 pKa = 8.95YY14 pKa = 10.78LDD16 pKa = 4.07RR17 pKa = 11.84DD18 pKa = 4.17SIQEE22 pKa = 4.09LNSLSHH28 pKa = 6.55NEE30 pKa = 4.66LMDD33 pKa = 3.55ILQRR37 pKa = 11.84LYY39 pKa = 10.17ISSDD43 pKa = 2.99KK44 pKa = 10.81EE45 pKa = 4.01YY46 pKa = 11.62VEE48 pKa = 4.22ITLSLSSSGGVNILFVDD65 pKa = 4.9DD66 pKa = 4.13YY67 pKa = 11.14PIVKK71 pKa = 9.75NQFITLYY78 pKa = 9.53NGEE81 pKa = 4.23VTDD84 pKa = 3.94EE85 pKa = 4.19SEE87 pKa = 4.25YY88 pKa = 11.62
MM1 pKa = 7.65NDD3 pKa = 3.16SEE5 pKa = 5.36VYY7 pKa = 8.19TFKK10 pKa = 10.77VRR12 pKa = 11.84YY13 pKa = 8.95YY14 pKa = 10.78LDD16 pKa = 4.07RR17 pKa = 11.84DD18 pKa = 4.17SIQEE22 pKa = 4.09LNSLSHH28 pKa = 6.55NEE30 pKa = 4.66LMDD33 pKa = 3.55ILQRR37 pKa = 11.84LYY39 pKa = 10.17ISSDD43 pKa = 2.99KK44 pKa = 10.81EE45 pKa = 4.01YY46 pKa = 11.62VEE48 pKa = 4.22ITLSLSSSGGVNILFVDD65 pKa = 4.9DD66 pKa = 4.13YY67 pKa = 11.14PIVKK71 pKa = 9.75NQFITLYY78 pKa = 9.53NGEE81 pKa = 4.23VTDD84 pKa = 3.94EE85 pKa = 4.19SEE87 pKa = 4.25YY88 pKa = 11.62
Molecular weight: 10.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A172Q064|A0A172Q064_9CAUD Putative exodeoxyribonuclease OS=Acinetobacter phage vB_AbaM_ME3 OX=1837876 GN=ME3_85 PE=4 SV=1
MM1 pKa = 7.98IDD3 pKa = 3.14NQGPIKK9 pKa = 9.9RR10 pKa = 11.84DD11 pKa = 3.04RR12 pKa = 11.84VQKK15 pKa = 10.28YY16 pKa = 8.47GRR18 pKa = 11.84KK19 pKa = 9.55SGLQEE24 pKa = 3.79AKK26 pKa = 10.6SFAEE30 pKa = 4.02RR31 pKa = 11.84NGMKK35 pKa = 9.52TLPLLTNKK43 pKa = 9.79HH44 pKa = 5.44KK45 pKa = 11.23NKK47 pKa = 10.31LYY49 pKa = 10.38RR50 pKa = 11.84KK51 pKa = 9.44NIEE54 pKa = 4.03RR55 pKa = 11.84NAKK58 pKa = 9.39SSSRR62 pKa = 11.84VIPPEE67 pKa = 3.42EE68 pKa = 3.96RR69 pKa = 11.84KK70 pKa = 10.09FKK72 pKa = 10.93LSTITGIGNQTSSVATPFLPSSRR95 pKa = 11.84IMNPPQPAKK104 pKa = 10.39SKK106 pKa = 8.41YY107 pKa = 8.26TSFLGDD113 pKa = 3.73MYY115 pKa = 11.65NKK117 pKa = 10.25LLSSFDD123 pKa = 3.48KK124 pKa = 10.88STDD127 pKa = 3.44QAIGGALGFTNRR139 pKa = 11.84NVVPGFTQNTSSGLSRR155 pKa = 11.84FDD157 pKa = 3.73VGNGDD162 pKa = 3.51PFKK165 pKa = 10.88YY166 pKa = 10.37SNRR169 pKa = 11.84FMKK172 pKa = 10.71GDD174 pKa = 4.08LDD176 pKa = 4.16DD177 pKa = 5.72LRR179 pKa = 11.84AAYY182 pKa = 9.61INEE185 pKa = 3.81AAKK188 pKa = 10.2GVRR191 pKa = 11.84VRR193 pKa = 11.84GDD195 pKa = 3.22NVNPFDD201 pKa = 3.87NSRR204 pKa = 11.84GARR207 pKa = 11.84NLGRR211 pKa = 11.84LRR213 pKa = 11.84QLQNRR218 pKa = 11.84INGGGQADD226 pKa = 5.67LIRR229 pKa = 11.84EE230 pKa = 4.37SNSIRR235 pKa = 11.84SQLDD239 pKa = 3.36DD240 pKa = 3.27VKK242 pKa = 11.01GRR244 pKa = 11.84QKK246 pKa = 11.06SIFPPRR252 pKa = 11.84NIGQGQSNYY261 pKa = 9.84EE262 pKa = 3.98SLVRR266 pKa = 11.84QQEE269 pKa = 4.39SLEE272 pKa = 3.87ARR274 pKa = 11.84QVVVNRR280 pKa = 11.84KK281 pKa = 8.6LEE283 pKa = 4.15RR284 pKa = 11.84NARR287 pKa = 11.84ILSTANEE294 pKa = 4.33YY295 pKa = 11.04SGATHH300 pKa = 7.12IKK302 pKa = 10.28GFLPKK307 pKa = 10.07EE308 pKa = 4.09GVGGYY313 pKa = 10.67GLGVSNYY320 pKa = 9.11LAASRR325 pKa = 11.84KK326 pKa = 8.1EE327 pKa = 3.85LFVRR331 pKa = 11.84GLGFTGGILGPSGAEE346 pKa = 3.24ALMNSFGIVTAKK358 pKa = 9.77QRR360 pKa = 11.84AVTQSKK366 pKa = 10.41GLLGKK371 pKa = 9.25MFSAGFQAPLMTYY384 pKa = 10.82GSLLFTASSGGDD396 pKa = 2.91LSDD399 pKa = 4.21FVEE402 pKa = 4.52TQLTYY407 pKa = 11.09AGGLQGWRR415 pKa = 11.84IGSSVGGMLTKK426 pKa = 10.14GTGANLSQIKK436 pKa = 8.97GGNMISAMRR445 pKa = 11.84AQGGRR450 pKa = 11.84AVLRR454 pKa = 11.84GAAGVVGGVTGFALGAGLVAGASWLAQDD482 pKa = 3.82ISSNRR487 pKa = 11.84STIRR491 pKa = 11.84KK492 pKa = 7.29IAKK495 pKa = 10.1DD496 pKa = 3.73FTTKK500 pKa = 9.41TATMNTQNTRR510 pKa = 11.84QSLTMRR516 pKa = 11.84QMAISKK522 pKa = 9.32LAKK525 pKa = 10.08SGLNDD530 pKa = 3.73RR531 pKa = 11.84ATLLGNEE538 pKa = 3.99SRR540 pKa = 11.84VLKK543 pKa = 10.97GLMM546 pKa = 3.76
MM1 pKa = 7.98IDD3 pKa = 3.14NQGPIKK9 pKa = 9.9RR10 pKa = 11.84DD11 pKa = 3.04RR12 pKa = 11.84VQKK15 pKa = 10.28YY16 pKa = 8.47GRR18 pKa = 11.84KK19 pKa = 9.55SGLQEE24 pKa = 3.79AKK26 pKa = 10.6SFAEE30 pKa = 4.02RR31 pKa = 11.84NGMKK35 pKa = 9.52TLPLLTNKK43 pKa = 9.79HH44 pKa = 5.44KK45 pKa = 11.23NKK47 pKa = 10.31LYY49 pKa = 10.38RR50 pKa = 11.84KK51 pKa = 9.44NIEE54 pKa = 4.03RR55 pKa = 11.84NAKK58 pKa = 9.39SSSRR62 pKa = 11.84VIPPEE67 pKa = 3.42EE68 pKa = 3.96RR69 pKa = 11.84KK70 pKa = 10.09FKK72 pKa = 10.93LSTITGIGNQTSSVATPFLPSSRR95 pKa = 11.84IMNPPQPAKK104 pKa = 10.39SKK106 pKa = 8.41YY107 pKa = 8.26TSFLGDD113 pKa = 3.73MYY115 pKa = 11.65NKK117 pKa = 10.25LLSSFDD123 pKa = 3.48KK124 pKa = 10.88STDD127 pKa = 3.44QAIGGALGFTNRR139 pKa = 11.84NVVPGFTQNTSSGLSRR155 pKa = 11.84FDD157 pKa = 3.73VGNGDD162 pKa = 3.51PFKK165 pKa = 10.88YY166 pKa = 10.37SNRR169 pKa = 11.84FMKK172 pKa = 10.71GDD174 pKa = 4.08LDD176 pKa = 4.16DD177 pKa = 5.72LRR179 pKa = 11.84AAYY182 pKa = 9.61INEE185 pKa = 3.81AAKK188 pKa = 10.2GVRR191 pKa = 11.84VRR193 pKa = 11.84GDD195 pKa = 3.22NVNPFDD201 pKa = 3.87NSRR204 pKa = 11.84GARR207 pKa = 11.84NLGRR211 pKa = 11.84LRR213 pKa = 11.84QLQNRR218 pKa = 11.84INGGGQADD226 pKa = 5.67LIRR229 pKa = 11.84EE230 pKa = 4.37SNSIRR235 pKa = 11.84SQLDD239 pKa = 3.36DD240 pKa = 3.27VKK242 pKa = 11.01GRR244 pKa = 11.84QKK246 pKa = 11.06SIFPPRR252 pKa = 11.84NIGQGQSNYY261 pKa = 9.84EE262 pKa = 3.98SLVRR266 pKa = 11.84QQEE269 pKa = 4.39SLEE272 pKa = 3.87ARR274 pKa = 11.84QVVVNRR280 pKa = 11.84KK281 pKa = 8.6LEE283 pKa = 4.15RR284 pKa = 11.84NARR287 pKa = 11.84ILSTANEE294 pKa = 4.33YY295 pKa = 11.04SGATHH300 pKa = 7.12IKK302 pKa = 10.28GFLPKK307 pKa = 10.07EE308 pKa = 4.09GVGGYY313 pKa = 10.67GLGVSNYY320 pKa = 9.11LAASRR325 pKa = 11.84KK326 pKa = 8.1EE327 pKa = 3.85LFVRR331 pKa = 11.84GLGFTGGILGPSGAEE346 pKa = 3.24ALMNSFGIVTAKK358 pKa = 9.77QRR360 pKa = 11.84AVTQSKK366 pKa = 10.41GLLGKK371 pKa = 9.25MFSAGFQAPLMTYY384 pKa = 10.82GSLLFTASSGGDD396 pKa = 2.91LSDD399 pKa = 4.21FVEE402 pKa = 4.52TQLTYY407 pKa = 11.09AGGLQGWRR415 pKa = 11.84IGSSVGGMLTKK426 pKa = 10.14GTGANLSQIKK436 pKa = 8.97GGNMISAMRR445 pKa = 11.84AQGGRR450 pKa = 11.84AVLRR454 pKa = 11.84GAAGVVGGVTGFALGAGLVAGASWLAQDD482 pKa = 3.82ISSNRR487 pKa = 11.84STIRR491 pKa = 11.84KK492 pKa = 7.29IAKK495 pKa = 10.1DD496 pKa = 3.73FTTKK500 pKa = 9.41TATMNTQNTRR510 pKa = 11.84QSLTMRR516 pKa = 11.84QMAISKK522 pKa = 9.32LAKK525 pKa = 10.08SGLNDD530 pKa = 3.73RR531 pKa = 11.84ATLLGNEE538 pKa = 3.99SRR540 pKa = 11.84VLKK543 pKa = 10.97GLMM546 pKa = 3.76
Molecular weight: 58.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
73381 |
27 |
5419 |
225.1 |
25.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.301 ± 0.31 | 1.055 ± 0.09 |
5.997 ± 0.106 | 6.119 ± 0.218 |
4.467 ± 0.121 | 5.024 ± 0.201 |
1.804 ± 0.106 | 7.675 ± 0.151 |
7.203 ± 0.211 | 9.212 ± 0.18 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.906 ± 0.08 | 6.517 ± 0.141 |
3.258 ± 0.074 | 3.355 ± 0.073 |
3.624 ± 0.101 | 8.246 ± 0.15 |
7.069 ± 0.35 | 6.485 ± 0.105 |
0.921 ± 0.052 | 4.76 ± 0.128 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
