
Bacillariodnavirus LDMD-2013
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Baphyvirales; Bacilladnaviridae; Protobacilladnavirus; Marine protobacilladnavirus 1
Average proteome isoelectric point is 7.27
Get precalculated fractions of proteins
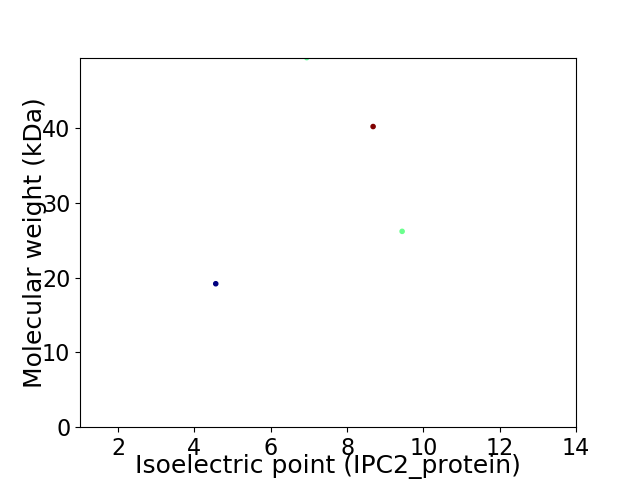
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
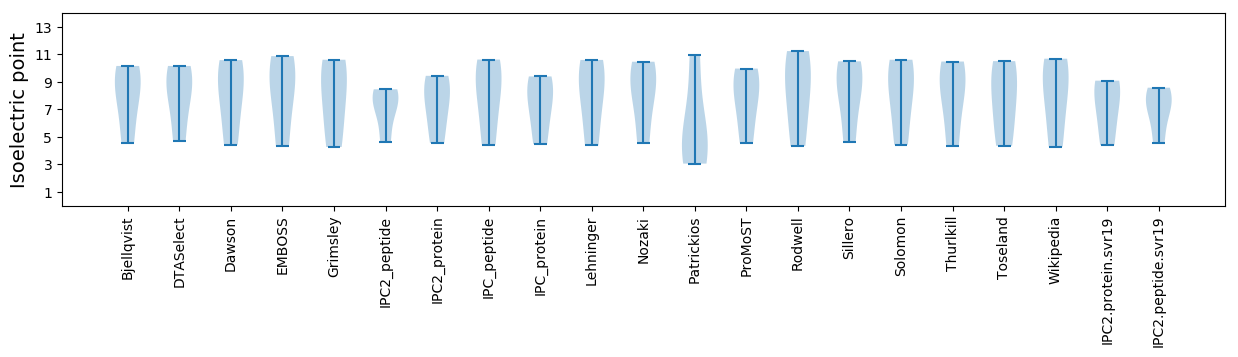
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5TMU4|S5TMU4_9VIRU Uncharacterized protein OS=Bacillariodnavirus LDMD-2013 OX=1379694 PE=4 SV=1
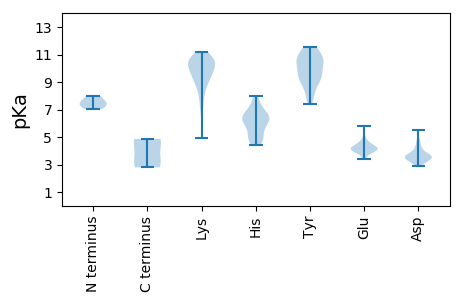
MM1 pKa = 7.45FGYY4 pKa = 9.94FFAILDD10 pKa = 3.76RR11 pKa = 11.84LDD13 pKa = 4.2WICHH17 pKa = 4.45FLFRR21 pKa = 11.84STMLEE26 pKa = 3.53RR27 pKa = 11.84TMVVLEE33 pKa = 4.21RR34 pKa = 11.84NGNRR38 pKa = 11.84VPYY41 pKa = 9.57DD42 pKa = 2.88IHH44 pKa = 6.5ATQCRR49 pKa = 11.84ICEE52 pKa = 3.97LWHH55 pKa = 5.86TLEE58 pKa = 4.46SAPEE62 pKa = 3.86EE63 pKa = 4.02EE64 pKa = 4.58RR65 pKa = 11.84MNRR68 pKa = 11.84AALEE72 pKa = 4.01LCNQKK77 pKa = 9.19IHH79 pKa = 6.63DD80 pKa = 4.41LKK82 pKa = 11.18FLVEE86 pKa = 4.0VDD88 pKa = 5.49HH89 pKa = 8.0DD90 pKa = 4.63DD91 pKa = 3.44FWPILINHH99 pKa = 6.65PNGLRR104 pKa = 11.84RR105 pKa = 11.84CTWVHH110 pKa = 4.76MFEE113 pKa = 3.59WWSIYY118 pKa = 10.07GVEE121 pKa = 4.58CVLEE125 pKa = 4.46EE126 pKa = 4.48IPIVDD131 pKa = 4.84NIQIAEE137 pKa = 4.09PLDD140 pKa = 3.77YY141 pKa = 11.58NEE143 pKa = 5.57DD144 pKa = 3.5GSIGTNTEE152 pKa = 4.06AEE154 pKa = 4.24MCDD157 pKa = 4.17LVAEE161 pKa = 4.59LL162 pKa = 4.87
MM1 pKa = 7.45FGYY4 pKa = 9.94FFAILDD10 pKa = 3.76RR11 pKa = 11.84LDD13 pKa = 4.2WICHH17 pKa = 4.45FLFRR21 pKa = 11.84STMLEE26 pKa = 3.53RR27 pKa = 11.84TMVVLEE33 pKa = 4.21RR34 pKa = 11.84NGNRR38 pKa = 11.84VPYY41 pKa = 9.57DD42 pKa = 2.88IHH44 pKa = 6.5ATQCRR49 pKa = 11.84ICEE52 pKa = 3.97LWHH55 pKa = 5.86TLEE58 pKa = 4.46SAPEE62 pKa = 3.86EE63 pKa = 4.02EE64 pKa = 4.58RR65 pKa = 11.84MNRR68 pKa = 11.84AALEE72 pKa = 4.01LCNQKK77 pKa = 9.19IHH79 pKa = 6.63DD80 pKa = 4.41LKK82 pKa = 11.18FLVEE86 pKa = 4.0VDD88 pKa = 5.49HH89 pKa = 8.0DD90 pKa = 4.63DD91 pKa = 3.44FWPILINHH99 pKa = 6.65PNGLRR104 pKa = 11.84RR105 pKa = 11.84CTWVHH110 pKa = 4.76MFEE113 pKa = 3.59WWSIYY118 pKa = 10.07GVEE121 pKa = 4.58CVLEE125 pKa = 4.46EE126 pKa = 4.48IPIVDD131 pKa = 4.84NIQIAEE137 pKa = 4.09PLDD140 pKa = 3.77YY141 pKa = 11.58NEE143 pKa = 5.57DD144 pKa = 3.5GSIGTNTEE152 pKa = 4.06AEE154 pKa = 4.24MCDD157 pKa = 4.17LVAEE161 pKa = 4.59LL162 pKa = 4.87
Molecular weight: 19.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5TNB3|S5TNB3_9VIRU Replication-associated protein OS=Bacillariodnavirus LDMD-2013 OX=1379694 PE=4 SV=1
MM1 pKa = 7.99DD2 pKa = 4.78IDD4 pKa = 3.61KK5 pKa = 10.34TSRR8 pKa = 11.84GVQSGIRR15 pKa = 11.84KK16 pKa = 9.19RR17 pKa = 11.84SNFEE21 pKa = 3.95EE22 pKa = 4.18IQNALQVVANKK33 pKa = 9.44KK34 pKa = 10.22AKK36 pKa = 10.06RR37 pKa = 11.84LDD39 pKa = 3.5YY40 pKa = 11.03AKK42 pKa = 10.39EE43 pKa = 3.9QGYY46 pKa = 9.53HH47 pKa = 6.52PLQLLGDD54 pKa = 3.97AVKK57 pKa = 10.75DD58 pKa = 3.32IGSVFLPEE66 pKa = 3.83QLASGVGGFITAGASEE82 pKa = 4.05PSIYY86 pKa = 10.32HH87 pKa = 5.86KK88 pKa = 10.86VGNVPTNVTEE98 pKa = 4.13KK99 pKa = 10.95DD100 pKa = 3.14AGQLLQSVSDD110 pKa = 3.39IYY112 pKa = 11.21EE113 pKa = 4.14GVGVLEE119 pKa = 4.52AEE121 pKa = 3.96QKK123 pKa = 9.92KK124 pKa = 10.06RR125 pKa = 11.84KK126 pKa = 9.08KK127 pKa = 10.73AEE129 pKa = 3.63AFIANEE135 pKa = 3.99ADD137 pKa = 3.3TSIGEE142 pKa = 4.27FEE144 pKa = 4.22SALAFLRR151 pKa = 11.84NEE153 pKa = 4.18VVPTGDD159 pKa = 3.24MDD161 pKa = 3.76QGEE164 pKa = 4.32LAEE167 pKa = 4.79AVPLPSGKK175 pKa = 9.77KK176 pKa = 9.92RR177 pKa = 11.84IVSKK181 pKa = 10.85KK182 pKa = 8.45KK183 pKa = 7.25TVKK186 pKa = 10.46RR187 pKa = 11.84KK188 pKa = 7.32ATKK191 pKa = 9.89PRR193 pKa = 11.84TTSKK197 pKa = 10.08RR198 pKa = 11.84RR199 pKa = 11.84KK200 pKa = 8.34VSPVKK205 pKa = 9.46KK206 pKa = 8.91TSGKK210 pKa = 9.47RR211 pKa = 11.84SKK213 pKa = 10.98KK214 pKa = 10.25SVTKK218 pKa = 10.59GSSKK222 pKa = 9.97RR223 pKa = 11.84RR224 pKa = 11.84KK225 pKa = 8.52GNYY228 pKa = 8.59RR229 pKa = 11.84VSPTGKK235 pKa = 9.16RR236 pKa = 11.84QRR238 pKa = 11.84VSS240 pKa = 2.8
MM1 pKa = 7.99DD2 pKa = 4.78IDD4 pKa = 3.61KK5 pKa = 10.34TSRR8 pKa = 11.84GVQSGIRR15 pKa = 11.84KK16 pKa = 9.19RR17 pKa = 11.84SNFEE21 pKa = 3.95EE22 pKa = 4.18IQNALQVVANKK33 pKa = 9.44KK34 pKa = 10.22AKK36 pKa = 10.06RR37 pKa = 11.84LDD39 pKa = 3.5YY40 pKa = 11.03AKK42 pKa = 10.39EE43 pKa = 3.9QGYY46 pKa = 9.53HH47 pKa = 6.52PLQLLGDD54 pKa = 3.97AVKK57 pKa = 10.75DD58 pKa = 3.32IGSVFLPEE66 pKa = 3.83QLASGVGGFITAGASEE82 pKa = 4.05PSIYY86 pKa = 10.32HH87 pKa = 5.86KK88 pKa = 10.86VGNVPTNVTEE98 pKa = 4.13KK99 pKa = 10.95DD100 pKa = 3.14AGQLLQSVSDD110 pKa = 3.39IYY112 pKa = 11.21EE113 pKa = 4.14GVGVLEE119 pKa = 4.52AEE121 pKa = 3.96QKK123 pKa = 9.92KK124 pKa = 10.06RR125 pKa = 11.84KK126 pKa = 9.08KK127 pKa = 10.73AEE129 pKa = 3.63AFIANEE135 pKa = 3.99ADD137 pKa = 3.3TSIGEE142 pKa = 4.27FEE144 pKa = 4.22SALAFLRR151 pKa = 11.84NEE153 pKa = 4.18VVPTGDD159 pKa = 3.24MDD161 pKa = 3.76QGEE164 pKa = 4.32LAEE167 pKa = 4.79AVPLPSGKK175 pKa = 9.77KK176 pKa = 9.92RR177 pKa = 11.84IVSKK181 pKa = 10.85KK182 pKa = 8.45KK183 pKa = 7.25TVKK186 pKa = 10.46RR187 pKa = 11.84KK188 pKa = 7.32ATKK191 pKa = 9.89PRR193 pKa = 11.84TTSKK197 pKa = 10.08RR198 pKa = 11.84RR199 pKa = 11.84KK200 pKa = 8.34VSPVKK205 pKa = 9.46KK206 pKa = 8.91TSGKK210 pKa = 9.47RR211 pKa = 11.84SKK213 pKa = 10.98KK214 pKa = 10.25SVTKK218 pKa = 10.59GSSKK222 pKa = 9.97RR223 pKa = 11.84RR224 pKa = 11.84KK225 pKa = 8.52GNYY228 pKa = 8.59RR229 pKa = 11.84VSPTGKK235 pKa = 9.16RR236 pKa = 11.84QRR238 pKa = 11.84VSS240 pKa = 2.8
Molecular weight: 26.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1191 |
162 |
423 |
297.8 |
33.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.213 ± 0.883 | 1.511 ± 0.588 |
5.374 ± 0.483 | 6.549 ± 0.934 |
4.282 ± 0.462 | 7.137 ± 0.897 |
3.61 ± 0.751 | 5.458 ± 0.482 |
6.801 ± 1.7 | 7.053 ± 0.638 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.511 ± 0.554 | 4.786 ± 0.431 |
4.45 ± 0.381 | 4.954 ± 0.679 |
6.297 ± 0.255 | 6.885 ± 0.839 |
6.213 ± 0.613 | 6.297 ± 0.748 |
2.183 ± 0.78 | 2.435 ± 0.153 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
