
Dishui Lake virophage 8
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Maveriviricetes; Priklausovirales; Lavidaviridae; unclassified Lavidaviridae
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
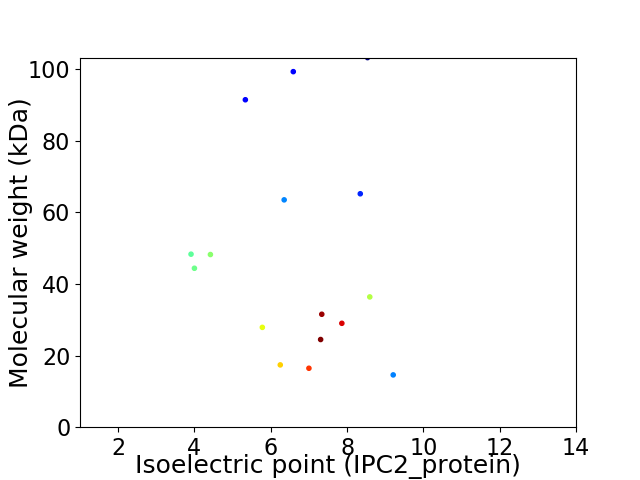
Virtual 2D-PAGE plot for 16 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
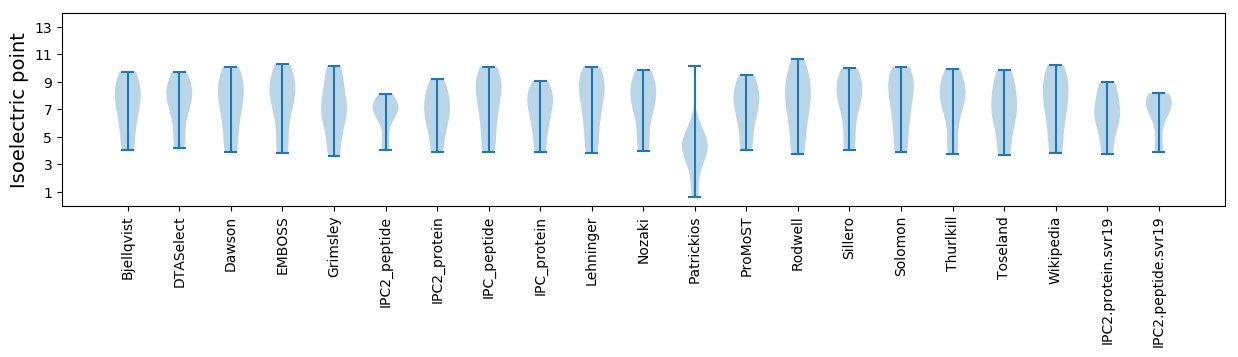
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G6XNK5|A0A6G6XNK5_9VIRU Putative collagen triple helix repeat-containing protein OS=Dishui Lake virophage 8 OX=2704069 PE=4 SV=1
MM1 pKa = 7.24SVSSIINPTDD11 pKa = 3.1GKK13 pKa = 10.73IYY15 pKa = 10.1TEE17 pKa = 4.89LYY19 pKa = 10.34NGGGGYY25 pKa = 10.37NLGEE29 pKa = 3.98VLSFGNNASNPTTGLPQDD47 pKa = 3.44ATDD50 pKa = 4.79FNNLGCISIEE60 pKa = 4.04TGKK63 pKa = 10.73VYY65 pKa = 10.54QGNNLDD71 pKa = 4.28LEE73 pKa = 4.49IGEE76 pKa = 5.19AGDD79 pKa = 3.42TLKK82 pKa = 10.75IIGAITKK89 pKa = 10.4GSILVGNGVEE99 pKa = 4.47TKK101 pKa = 9.82EE102 pKa = 4.51LPVGANGLVLKK113 pKa = 10.48ANSAATYY120 pKa = 8.56GVEE123 pKa = 3.88WAVDD127 pKa = 3.6GTTGITGVSAGTNIGIDD144 pKa = 3.46NTNPIVPIINFATPTTSNISLGVGTFIEE172 pKa = 4.73AKK174 pKa = 10.39DD175 pKa = 3.78NYY177 pKa = 8.94STPNFAMTLDD187 pKa = 3.36ATGFNDD193 pKa = 3.93TYY195 pKa = 11.35TNGAVEE201 pKa = 4.33NKK203 pKa = 10.2EE204 pKa = 4.21DD205 pKa = 3.72IEE207 pKa = 4.62VNATSVVQLLSATDD221 pKa = 3.37GGSYY225 pKa = 10.98LNTDD229 pKa = 3.5TTNVSNTGIVEE240 pKa = 4.11NLSATNLSNNNIGNVSFTCYY260 pKa = 10.51SNSAGLACGCSAPTTPPYY278 pKa = 10.56PEE280 pKa = 4.09VSATAGLSATTTNAQLTISQSAPFAISYY308 pKa = 7.62STILDD313 pKa = 3.33INGINQNNTGGVAGFTINTNTQPLALTTGDD343 pKa = 4.42NITFSADD350 pKa = 3.85NIDD353 pKa = 3.92LDD355 pKa = 3.72ATGRR359 pKa = 11.84LILPSLASGDD369 pKa = 3.84YY370 pKa = 10.74LDD372 pKa = 5.15YY373 pKa = 11.34NAGSLKK379 pKa = 10.2IVNDD383 pKa = 3.85SVAGSANPLLVLQNNNNTAGATTFEE408 pKa = 4.78TYY410 pKa = 10.85KK411 pKa = 10.65NDD413 pKa = 3.34QPTSTGGDD421 pKa = 3.72NIASWSATCNTNVGKK436 pKa = 8.65TEE438 pKa = 3.64ISRR441 pKa = 11.84INHH444 pKa = 5.69IALLTEE450 pKa = 4.33LVLLIMTAVLPLLVRR465 pKa = 11.84LIRR468 pKa = 11.84QQ469 pKa = 3.25
MM1 pKa = 7.24SVSSIINPTDD11 pKa = 3.1GKK13 pKa = 10.73IYY15 pKa = 10.1TEE17 pKa = 4.89LYY19 pKa = 10.34NGGGGYY25 pKa = 10.37NLGEE29 pKa = 3.98VLSFGNNASNPTTGLPQDD47 pKa = 3.44ATDD50 pKa = 4.79FNNLGCISIEE60 pKa = 4.04TGKK63 pKa = 10.73VYY65 pKa = 10.54QGNNLDD71 pKa = 4.28LEE73 pKa = 4.49IGEE76 pKa = 5.19AGDD79 pKa = 3.42TLKK82 pKa = 10.75IIGAITKK89 pKa = 10.4GSILVGNGVEE99 pKa = 4.47TKK101 pKa = 9.82EE102 pKa = 4.51LPVGANGLVLKK113 pKa = 10.48ANSAATYY120 pKa = 8.56GVEE123 pKa = 3.88WAVDD127 pKa = 3.6GTTGITGVSAGTNIGIDD144 pKa = 3.46NTNPIVPIINFATPTTSNISLGVGTFIEE172 pKa = 4.73AKK174 pKa = 10.39DD175 pKa = 3.78NYY177 pKa = 8.94STPNFAMTLDD187 pKa = 3.36ATGFNDD193 pKa = 3.93TYY195 pKa = 11.35TNGAVEE201 pKa = 4.33NKK203 pKa = 10.2EE204 pKa = 4.21DD205 pKa = 3.72IEE207 pKa = 4.62VNATSVVQLLSATDD221 pKa = 3.37GGSYY225 pKa = 10.98LNTDD229 pKa = 3.5TTNVSNTGIVEE240 pKa = 4.11NLSATNLSNNNIGNVSFTCYY260 pKa = 10.51SNSAGLACGCSAPTTPPYY278 pKa = 10.56PEE280 pKa = 4.09VSATAGLSATTTNAQLTISQSAPFAISYY308 pKa = 7.62STILDD313 pKa = 3.33INGINQNNTGGVAGFTINTNTQPLALTTGDD343 pKa = 4.42NITFSADD350 pKa = 3.85NIDD353 pKa = 3.92LDD355 pKa = 3.72ATGRR359 pKa = 11.84LILPSLASGDD369 pKa = 3.84YY370 pKa = 10.74LDD372 pKa = 5.15YY373 pKa = 11.34NAGSLKK379 pKa = 10.2IVNDD383 pKa = 3.85SVAGSANPLLVLQNNNNTAGATTFEE408 pKa = 4.78TYY410 pKa = 10.85KK411 pKa = 10.65NDD413 pKa = 3.34QPTSTGGDD421 pKa = 3.72NIASWSATCNTNVGKK436 pKa = 8.65TEE438 pKa = 3.64ISRR441 pKa = 11.84INHH444 pKa = 5.69IALLTEE450 pKa = 4.33LVLLIMTAVLPLLVRR465 pKa = 11.84LIRR468 pKa = 11.84QQ469 pKa = 3.25
Molecular weight: 48.33 kDa
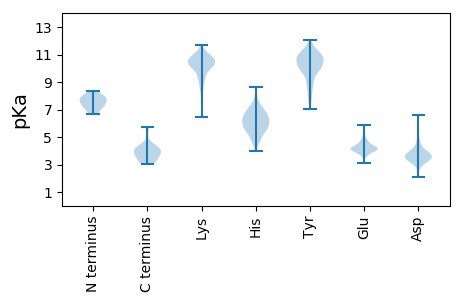
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G6XMW4|A0A6G6XMW4_9VIRU Putative major capsid protein OS=Dishui Lake virophage 8 OX=2704069 PE=4 SV=1
MM1 pKa = 7.28NVFNHH6 pKa = 7.17FIYY9 pKa = 10.2IIYY12 pKa = 8.94IDD14 pKa = 4.38KK15 pKa = 10.35MFSNQLKK22 pKa = 9.43EE23 pKa = 3.93LKK25 pKa = 10.32IPIKK29 pKa = 10.06TYY31 pKa = 11.18LNTAKK36 pKa = 10.02QRR38 pKa = 11.84AKK40 pKa = 10.68NAGYY44 pKa = 10.1DD45 pKa = 3.7PKK47 pKa = 10.99LLSLSKK53 pKa = 10.9DD54 pKa = 3.07KK55 pKa = 10.73EE56 pKa = 4.14YY57 pKa = 11.25KK58 pKa = 10.44LNYY61 pKa = 9.8DD62 pKa = 3.35GVNFGRR68 pKa = 11.84TGYY71 pKa = 10.95GDD73 pKa = 4.09FIIWSILEE81 pKa = 3.88DD82 pKa = 3.29RR83 pKa = 11.84GLVEE87 pKa = 5.62KK88 pKa = 10.87GYY90 pKa = 11.25ADD92 pKa = 3.23KK93 pKa = 10.84KK94 pKa = 9.79QNVFRR99 pKa = 11.84ASHH102 pKa = 5.83SKK104 pKa = 10.47IKK106 pKa = 10.71GDD108 pKa = 3.23WKK110 pKa = 11.11NNPKK114 pKa = 10.34SPNNLALKK122 pKa = 10.4INWW125 pKa = 3.44
MM1 pKa = 7.28NVFNHH6 pKa = 7.17FIYY9 pKa = 10.2IIYY12 pKa = 8.94IDD14 pKa = 4.38KK15 pKa = 10.35MFSNQLKK22 pKa = 9.43EE23 pKa = 3.93LKK25 pKa = 10.32IPIKK29 pKa = 10.06TYY31 pKa = 11.18LNTAKK36 pKa = 10.02QRR38 pKa = 11.84AKK40 pKa = 10.68NAGYY44 pKa = 10.1DD45 pKa = 3.7PKK47 pKa = 10.99LLSLSKK53 pKa = 10.9DD54 pKa = 3.07KK55 pKa = 10.73EE56 pKa = 4.14YY57 pKa = 11.25KK58 pKa = 10.44LNYY61 pKa = 9.8DD62 pKa = 3.35GVNFGRR68 pKa = 11.84TGYY71 pKa = 10.95GDD73 pKa = 4.09FIIWSILEE81 pKa = 3.88DD82 pKa = 3.29RR83 pKa = 11.84GLVEE87 pKa = 5.62KK88 pKa = 10.87GYY90 pKa = 11.25ADD92 pKa = 3.23KK93 pKa = 10.84KK94 pKa = 9.79QNVFRR99 pKa = 11.84ASHH102 pKa = 5.83SKK104 pKa = 10.47IKK106 pKa = 10.71GDD108 pKa = 3.23WKK110 pKa = 11.11NNPKK114 pKa = 10.34SPNNLALKK122 pKa = 10.4INWW125 pKa = 3.44
Molecular weight: 14.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6793 |
125 |
890 |
424.6 |
47.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.668 ± 0.681 | 0.972 ± 0.18 |
6.198 ± 0.295 | 5.491 ± 0.701 |
3.842 ± 0.293 | 7.272 ± 1.098 |
1.104 ± 0.203 | 7.169 ± 0.474 |
8.008 ± 1.261 | 8.052 ± 0.314 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.267 ± 0.305 | 7.316 ± 0.659 |
4.887 ± 0.432 | 4.431 ± 0.687 |
3.562 ± 0.489 | 6.58 ± 0.658 |
6.242 ± 0.725 | 5.182 ± 0.334 |
1.075 ± 0.145 | 4.608 ± 0.405 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
