
Theileria parva (East coast fever infection agent)
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Aconoidasida; Piroplasmida; Theileriidae; Theileria
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
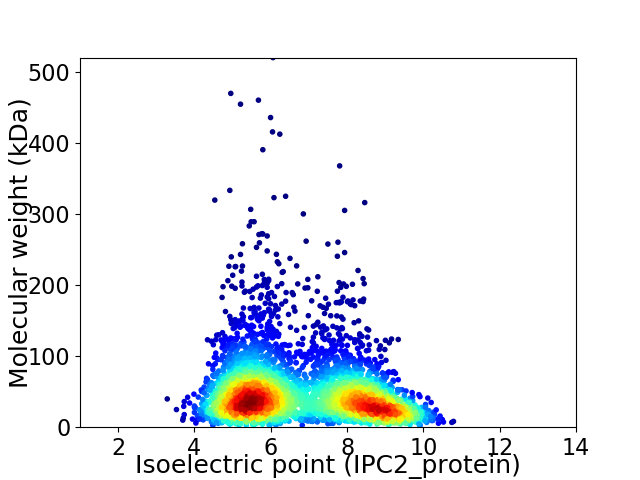
Virtual 2D-PAGE plot for 4071 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q4MZP6|Q4MZP6_THEPA Uncharacterized protein OS=Theileria parva OX=5875 GN=TP03_0471 PE=4 SV=1
MM1 pKa = 7.7SSTILNSINSLDD13 pKa = 3.58YY14 pKa = 10.85SLEE17 pKa = 4.04VEE19 pKa = 4.07SDD21 pKa = 3.65GNFGKK26 pKa = 8.72FTDD29 pKa = 3.54QLVYY33 pKa = 10.62EE34 pKa = 4.39VVKK37 pKa = 10.98LSEE40 pKa = 4.29CDD42 pKa = 3.48YY43 pKa = 11.44DD44 pKa = 4.36PEE46 pKa = 4.6TEE48 pKa = 4.07TFYY51 pKa = 11.22YY52 pKa = 10.62LCPCGDD58 pKa = 3.78LFEE61 pKa = 5.14IALEE65 pKa = 4.34DD66 pKa = 3.91LLKK69 pKa = 11.24GNLISEE75 pKa = 4.83CPSCSLRR82 pKa = 11.84IRR84 pKa = 11.84IDD86 pKa = 3.88LKK88 pKa = 11.07PGDD91 pKa = 4.28LDD93 pKa = 3.39QFVNMKK99 pKa = 8.9TVV101 pKa = 2.66
MM1 pKa = 7.7SSTILNSINSLDD13 pKa = 3.58YY14 pKa = 10.85SLEE17 pKa = 4.04VEE19 pKa = 4.07SDD21 pKa = 3.65GNFGKK26 pKa = 8.72FTDD29 pKa = 3.54QLVYY33 pKa = 10.62EE34 pKa = 4.39VVKK37 pKa = 10.98LSEE40 pKa = 4.29CDD42 pKa = 3.48YY43 pKa = 11.44DD44 pKa = 4.36PEE46 pKa = 4.6TEE48 pKa = 4.07TFYY51 pKa = 11.22YY52 pKa = 10.62LCPCGDD58 pKa = 3.78LFEE61 pKa = 5.14IALEE65 pKa = 4.34DD66 pKa = 3.91LLKK69 pKa = 11.24GNLISEE75 pKa = 4.83CPSCSLRR82 pKa = 11.84IRR84 pKa = 11.84IDD86 pKa = 3.88LKK88 pKa = 11.07PGDD91 pKa = 4.28LDD93 pKa = 3.39QFVNMKK99 pKa = 8.9TVV101 pKa = 2.66
Molecular weight: 11.42 kDa
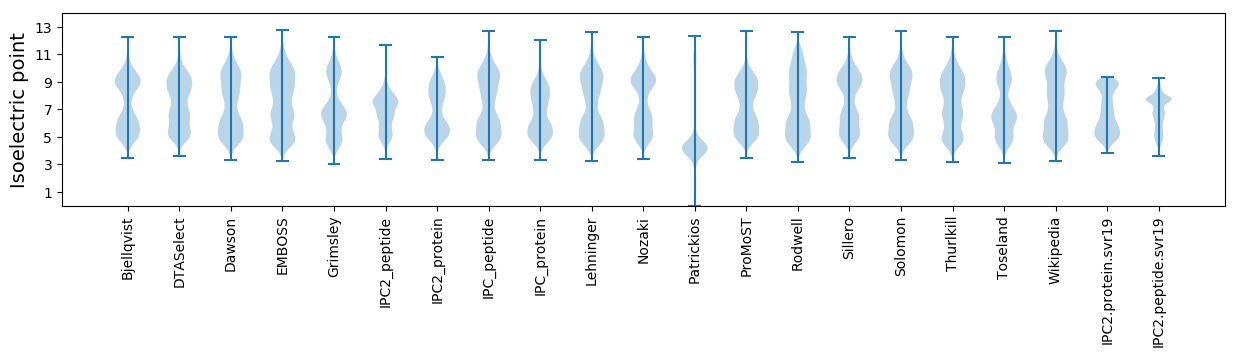
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q4N2T1|Q4N2T1_THEPA Small nuclear ribonucleoprotein putative OS=Theileria parva OX=5875 GN=TP04_0265 PE=3 SV=1
MM1 pKa = 7.74GMDD4 pKa = 4.12DD5 pKa = 5.05LSRR8 pKa = 11.84IYY10 pKa = 10.21IGNLPEE16 pKa = 5.1DD17 pKa = 3.93CSQRR21 pKa = 11.84EE22 pKa = 4.09LEE24 pKa = 4.23EE25 pKa = 3.98EE26 pKa = 4.05FEE28 pKa = 4.24KK29 pKa = 10.71FGRR32 pKa = 11.84IIYY35 pKa = 10.26CEE37 pKa = 4.0LKK39 pKa = 10.37KK40 pKa = 10.79SYY42 pKa = 10.28SGSPFAFIEE51 pKa = 4.63FSDD54 pKa = 4.05SRR56 pKa = 11.84DD57 pKa = 3.31ARR59 pKa = 11.84DD60 pKa = 4.03AIRR63 pKa = 11.84DD64 pKa = 3.25KK65 pKa = 11.21DD66 pKa = 3.57GYY68 pKa = 10.0EE69 pKa = 3.77FHH71 pKa = 6.77GKK73 pKa = 9.25KK74 pKa = 10.27LRR76 pKa = 11.84VEE78 pKa = 3.89LPFRR82 pKa = 11.84YY83 pKa = 8.99KK84 pKa = 10.73DD85 pKa = 3.44EE86 pKa = 4.09PRR88 pKa = 11.84RR89 pKa = 11.84PSGRR93 pKa = 11.84RR94 pKa = 11.84YY95 pKa = 8.06RR96 pKa = 11.84TTRR99 pKa = 11.84RR100 pKa = 11.84GKK102 pKa = 10.12YY103 pKa = 8.06VLEE106 pKa = 4.31VTGLPPSGSWQDD118 pKa = 4.13LKK120 pKa = 11.74DD121 pKa = 3.54HH122 pKa = 6.3MRR124 pKa = 11.84DD125 pKa = 3.3AGEE128 pKa = 4.73CGHH131 pKa = 7.15ADD133 pKa = 3.49VFRR136 pKa = 11.84GGVGEE141 pKa = 3.83ITFFSRR147 pKa = 11.84SDD149 pKa = 2.89MDD151 pKa = 3.71YY152 pKa = 10.86AIEE155 pKa = 4.4RR156 pKa = 11.84FDD158 pKa = 3.48GSTFRR163 pKa = 11.84SHH165 pKa = 6.75EE166 pKa = 4.25GEE168 pKa = 3.66KK169 pKa = 10.62SRR171 pKa = 11.84ISVRR175 pKa = 11.84EE176 pKa = 3.34KK177 pKa = 9.34SRR179 pKa = 11.84RR180 pKa = 11.84RR181 pKa = 11.84RR182 pKa = 11.84TRR184 pKa = 11.84SYY186 pKa = 10.44SRR188 pKa = 11.84DD189 pKa = 3.07RR190 pKa = 11.84GRR192 pKa = 11.84SYY194 pKa = 11.17SRR196 pKa = 11.84NNRR199 pKa = 11.84RR200 pKa = 11.84SRR202 pKa = 11.84SYY204 pKa = 9.99SRR206 pKa = 11.84SRR208 pKa = 11.84TRR210 pKa = 11.84SRR212 pKa = 11.84SRR214 pKa = 11.84TRR216 pKa = 11.84DD217 pKa = 2.92RR218 pKa = 11.84SRR220 pKa = 11.84DD221 pKa = 3.22RR222 pKa = 11.84SRR224 pKa = 11.84SRR226 pKa = 11.84DD227 pKa = 3.28RR228 pKa = 11.84NRR230 pKa = 11.84DD231 pKa = 3.3RR232 pKa = 11.84TSSRR236 pKa = 11.84SYY238 pKa = 11.31SRR240 pKa = 11.84DD241 pKa = 3.1KK242 pKa = 11.09SDD244 pKa = 3.29SRR246 pKa = 11.84SRR248 pKa = 11.84DD249 pKa = 3.2KK250 pKa = 10.79TVSRR254 pKa = 11.84SRR256 pKa = 11.84SS257 pKa = 3.31
MM1 pKa = 7.74GMDD4 pKa = 4.12DD5 pKa = 5.05LSRR8 pKa = 11.84IYY10 pKa = 10.21IGNLPEE16 pKa = 5.1DD17 pKa = 3.93CSQRR21 pKa = 11.84EE22 pKa = 4.09LEE24 pKa = 4.23EE25 pKa = 3.98EE26 pKa = 4.05FEE28 pKa = 4.24KK29 pKa = 10.71FGRR32 pKa = 11.84IIYY35 pKa = 10.26CEE37 pKa = 4.0LKK39 pKa = 10.37KK40 pKa = 10.79SYY42 pKa = 10.28SGSPFAFIEE51 pKa = 4.63FSDD54 pKa = 4.05SRR56 pKa = 11.84DD57 pKa = 3.31ARR59 pKa = 11.84DD60 pKa = 4.03AIRR63 pKa = 11.84DD64 pKa = 3.25KK65 pKa = 11.21DD66 pKa = 3.57GYY68 pKa = 10.0EE69 pKa = 3.77FHH71 pKa = 6.77GKK73 pKa = 9.25KK74 pKa = 10.27LRR76 pKa = 11.84VEE78 pKa = 3.89LPFRR82 pKa = 11.84YY83 pKa = 8.99KK84 pKa = 10.73DD85 pKa = 3.44EE86 pKa = 4.09PRR88 pKa = 11.84RR89 pKa = 11.84PSGRR93 pKa = 11.84RR94 pKa = 11.84YY95 pKa = 8.06RR96 pKa = 11.84TTRR99 pKa = 11.84RR100 pKa = 11.84GKK102 pKa = 10.12YY103 pKa = 8.06VLEE106 pKa = 4.31VTGLPPSGSWQDD118 pKa = 4.13LKK120 pKa = 11.74DD121 pKa = 3.54HH122 pKa = 6.3MRR124 pKa = 11.84DD125 pKa = 3.3AGEE128 pKa = 4.73CGHH131 pKa = 7.15ADD133 pKa = 3.49VFRR136 pKa = 11.84GGVGEE141 pKa = 3.83ITFFSRR147 pKa = 11.84SDD149 pKa = 2.89MDD151 pKa = 3.71YY152 pKa = 10.86AIEE155 pKa = 4.4RR156 pKa = 11.84FDD158 pKa = 3.48GSTFRR163 pKa = 11.84SHH165 pKa = 6.75EE166 pKa = 4.25GEE168 pKa = 3.66KK169 pKa = 10.62SRR171 pKa = 11.84ISVRR175 pKa = 11.84EE176 pKa = 3.34KK177 pKa = 9.34SRR179 pKa = 11.84RR180 pKa = 11.84RR181 pKa = 11.84RR182 pKa = 11.84TRR184 pKa = 11.84SYY186 pKa = 10.44SRR188 pKa = 11.84DD189 pKa = 3.07RR190 pKa = 11.84GRR192 pKa = 11.84SYY194 pKa = 11.17SRR196 pKa = 11.84NNRR199 pKa = 11.84RR200 pKa = 11.84SRR202 pKa = 11.84SYY204 pKa = 9.99SRR206 pKa = 11.84SRR208 pKa = 11.84TRR210 pKa = 11.84SRR212 pKa = 11.84SRR214 pKa = 11.84TRR216 pKa = 11.84DD217 pKa = 2.92RR218 pKa = 11.84SRR220 pKa = 11.84DD221 pKa = 3.22RR222 pKa = 11.84SRR224 pKa = 11.84SRR226 pKa = 11.84DD227 pKa = 3.28RR228 pKa = 11.84NRR230 pKa = 11.84DD231 pKa = 3.3RR232 pKa = 11.84TSSRR236 pKa = 11.84SYY238 pKa = 11.31SRR240 pKa = 11.84DD241 pKa = 3.1KK242 pKa = 11.09SDD244 pKa = 3.29SRR246 pKa = 11.84SRR248 pKa = 11.84DD249 pKa = 3.2KK250 pKa = 10.79TVSRR254 pKa = 11.84SRR256 pKa = 11.84SS257 pKa = 3.31
Molecular weight: 30.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1895349 |
19 |
4593 |
465.6 |
53.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.421 ± 0.034 | 1.661 ± 0.017 |
5.872 ± 0.032 | 6.368 ± 0.041 |
4.953 ± 0.028 | 4.469 ± 0.03 |
2.16 ± 0.015 | 6.565 ± 0.043 |
7.829 ± 0.042 | 10.186 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.033 ± 0.017 | 7.262 ± 0.05 |
3.946 ± 0.032 | 3.194 ± 0.033 |
4.139 ± 0.026 | 8.725 ± 0.042 |
5.901 ± 0.036 | 6.235 ± 0.029 |
0.795 ± 0.009 | 4.286 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
