
Changjiang crawfish virus 7
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.62
Get precalculated fractions of proteins
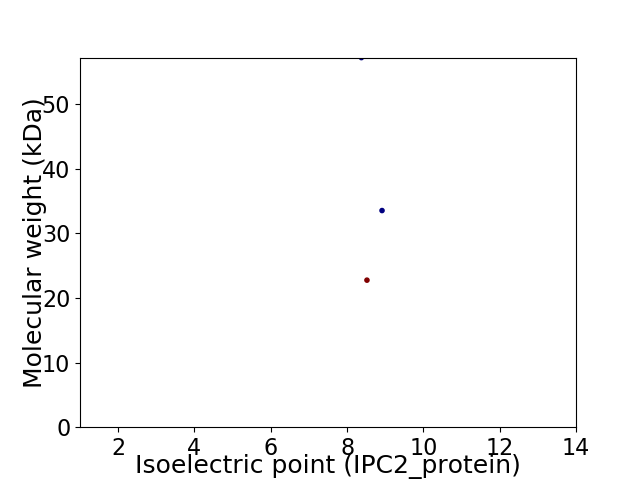
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFS4|A0A1L3KFS4_9VIRU Uncharacterized protein OS=Changjiang crawfish virus 7 OX=1922771 PE=4 SV=1
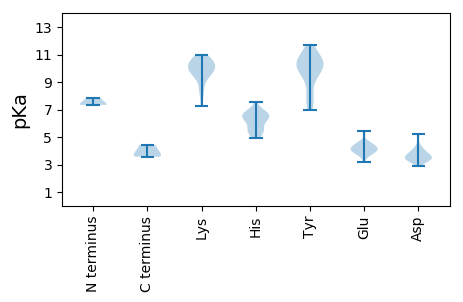
MM1 pKa = 7.84PEE3 pKa = 3.8VSVDD7 pKa = 3.45QQGYY11 pKa = 7.01SQKK14 pKa = 10.5LPKK17 pKa = 10.07RR18 pKa = 11.84LFDD21 pKa = 3.57IRR23 pKa = 11.84GLPGRR28 pKa = 11.84GTLWTHH34 pKa = 6.43ANDD37 pKa = 3.28VANVVQSVHH46 pKa = 5.44EE47 pKa = 4.18RR48 pKa = 11.84VLGRR52 pKa = 11.84TTSKK56 pKa = 10.0GWEE59 pKa = 3.79RR60 pKa = 11.84TLLPEE65 pKa = 4.22EE66 pKa = 4.52GAFDD70 pKa = 3.64SEE72 pKa = 4.32GMVRR76 pKa = 11.84FSTRR80 pKa = 11.84LKK82 pKa = 10.47QKK84 pKa = 10.32LGSHH88 pKa = 6.26SLPVTEE94 pKa = 5.45QDD96 pKa = 3.4FLSHH100 pKa = 6.14YY101 pKa = 9.81RR102 pKa = 11.84GQKK105 pKa = 7.76RR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84YY109 pKa = 7.91EE110 pKa = 3.7AAVSSLRR117 pKa = 11.84AKK119 pKa = 9.86PVQRR123 pKa = 11.84ADD125 pKa = 3.38SYY127 pKa = 11.63PSVFLKK133 pKa = 10.66AEE135 pKa = 3.97KK136 pKa = 9.52WRR138 pKa = 11.84EE139 pKa = 3.96EE140 pKa = 3.95KK141 pKa = 10.32PGRR144 pKa = 11.84LISARR149 pKa = 11.84SPRR152 pKa = 11.84YY153 pKa = 7.9NVEE156 pKa = 3.54VGRR159 pKa = 11.84YY160 pKa = 8.15LLPLEE165 pKa = 4.31PLVYY169 pKa = 10.1QAIDD173 pKa = 4.12GVWGSATIMKK183 pKa = 10.2GYY185 pKa = 8.24TPEE188 pKa = 3.47RR189 pKa = 11.84RR190 pKa = 11.84AAVVRR195 pKa = 11.84GHH197 pKa = 6.85WDD199 pKa = 3.14SFGDD203 pKa = 3.97PVAVGHH209 pKa = 6.79DD210 pKa = 3.69FSKK213 pKa = 10.84FDD215 pKa = 3.2QHH217 pKa = 6.7ISKK220 pKa = 10.6RR221 pKa = 11.84ALQYY225 pKa = 10.07EE226 pKa = 4.13HH227 pKa = 6.69GVYY230 pKa = 10.36LRR232 pKa = 11.84AYY234 pKa = 10.05AGDD237 pKa = 3.49EE238 pKa = 4.08HH239 pKa = 6.77LQKK242 pKa = 10.93LLSWQLEE249 pKa = 4.57TTCYY253 pKa = 11.14ANVRR257 pKa = 11.84DD258 pKa = 4.02GRR260 pKa = 11.84VKK262 pKa = 9.57YY263 pKa = 7.51TVKK266 pKa = 10.23GGRR269 pKa = 11.84MSGDD273 pKa = 3.27MNTAMGNCIISAGLIWAYY291 pKa = 10.2AAEE294 pKa = 4.41SGVQLRR300 pKa = 11.84AVVDD304 pKa = 3.91GDD306 pKa = 3.82DD307 pKa = 3.43SVVFMEE313 pKa = 5.34KK314 pKa = 10.1KK315 pKa = 10.33DD316 pKa = 3.69LQRR319 pKa = 11.84YY320 pKa = 7.31LAGIEE325 pKa = 3.5GWMAKK330 pKa = 9.98RR331 pKa = 11.84GFRR334 pKa = 11.84LVTEE338 pKa = 4.27EE339 pKa = 3.99PVYY342 pKa = 10.42EE343 pKa = 4.03INRR346 pKa = 11.84VEE348 pKa = 4.3FCQCRR353 pKa = 11.84YY354 pKa = 9.22MDD356 pKa = 4.21TVPPTMVRR364 pKa = 11.84NPLKK368 pKa = 10.77AITQDD373 pKa = 3.48HH374 pKa = 6.24AWIEE378 pKa = 4.23DD379 pKa = 3.41RR380 pKa = 11.84SISYY384 pKa = 11.09AEE386 pKa = 3.91VLAATGLGGLSLYY399 pKa = 10.74GHH401 pKa = 6.79IPVLGAYY408 pKa = 9.76YY409 pKa = 10.7DD410 pKa = 3.77LLARR414 pKa = 11.84TTQPSRR420 pKa = 11.84RR421 pKa = 11.84TLNRR425 pKa = 11.84LDD427 pKa = 5.17FRR429 pKa = 11.84SSWLRR434 pKa = 11.84DD435 pKa = 3.04ATMSGGFTEE444 pKa = 4.66PSEE447 pKa = 3.92QARR450 pKa = 11.84YY451 pKa = 9.36QFWLTWGMSPGEE463 pKa = 4.08QRR465 pKa = 11.84AHH467 pKa = 5.49EE468 pKa = 4.27MNFRR472 pKa = 11.84ACDD475 pKa = 3.76LQDD478 pKa = 4.17LVASDD483 pKa = 5.2TITKK487 pKa = 8.4ATRR490 pKa = 11.84NEE492 pKa = 3.82KK493 pKa = 9.31TDD495 pKa = 3.64PYY497 pKa = 10.47GNNYY501 pKa = 9.37YY502 pKa = 10.77LL503 pKa = 4.42
MM1 pKa = 7.84PEE3 pKa = 3.8VSVDD7 pKa = 3.45QQGYY11 pKa = 7.01SQKK14 pKa = 10.5LPKK17 pKa = 10.07RR18 pKa = 11.84LFDD21 pKa = 3.57IRR23 pKa = 11.84GLPGRR28 pKa = 11.84GTLWTHH34 pKa = 6.43ANDD37 pKa = 3.28VANVVQSVHH46 pKa = 5.44EE47 pKa = 4.18RR48 pKa = 11.84VLGRR52 pKa = 11.84TTSKK56 pKa = 10.0GWEE59 pKa = 3.79RR60 pKa = 11.84TLLPEE65 pKa = 4.22EE66 pKa = 4.52GAFDD70 pKa = 3.64SEE72 pKa = 4.32GMVRR76 pKa = 11.84FSTRR80 pKa = 11.84LKK82 pKa = 10.47QKK84 pKa = 10.32LGSHH88 pKa = 6.26SLPVTEE94 pKa = 5.45QDD96 pKa = 3.4FLSHH100 pKa = 6.14YY101 pKa = 9.81RR102 pKa = 11.84GQKK105 pKa = 7.76RR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84YY109 pKa = 7.91EE110 pKa = 3.7AAVSSLRR117 pKa = 11.84AKK119 pKa = 9.86PVQRR123 pKa = 11.84ADD125 pKa = 3.38SYY127 pKa = 11.63PSVFLKK133 pKa = 10.66AEE135 pKa = 3.97KK136 pKa = 9.52WRR138 pKa = 11.84EE139 pKa = 3.96EE140 pKa = 3.95KK141 pKa = 10.32PGRR144 pKa = 11.84LISARR149 pKa = 11.84SPRR152 pKa = 11.84YY153 pKa = 7.9NVEE156 pKa = 3.54VGRR159 pKa = 11.84YY160 pKa = 8.15LLPLEE165 pKa = 4.31PLVYY169 pKa = 10.1QAIDD173 pKa = 4.12GVWGSATIMKK183 pKa = 10.2GYY185 pKa = 8.24TPEE188 pKa = 3.47RR189 pKa = 11.84RR190 pKa = 11.84AAVVRR195 pKa = 11.84GHH197 pKa = 6.85WDD199 pKa = 3.14SFGDD203 pKa = 3.97PVAVGHH209 pKa = 6.79DD210 pKa = 3.69FSKK213 pKa = 10.84FDD215 pKa = 3.2QHH217 pKa = 6.7ISKK220 pKa = 10.6RR221 pKa = 11.84ALQYY225 pKa = 10.07EE226 pKa = 4.13HH227 pKa = 6.69GVYY230 pKa = 10.36LRR232 pKa = 11.84AYY234 pKa = 10.05AGDD237 pKa = 3.49EE238 pKa = 4.08HH239 pKa = 6.77LQKK242 pKa = 10.93LLSWQLEE249 pKa = 4.57TTCYY253 pKa = 11.14ANVRR257 pKa = 11.84DD258 pKa = 4.02GRR260 pKa = 11.84VKK262 pKa = 9.57YY263 pKa = 7.51TVKK266 pKa = 10.23GGRR269 pKa = 11.84MSGDD273 pKa = 3.27MNTAMGNCIISAGLIWAYY291 pKa = 10.2AAEE294 pKa = 4.41SGVQLRR300 pKa = 11.84AVVDD304 pKa = 3.91GDD306 pKa = 3.82DD307 pKa = 3.43SVVFMEE313 pKa = 5.34KK314 pKa = 10.1KK315 pKa = 10.33DD316 pKa = 3.69LQRR319 pKa = 11.84YY320 pKa = 7.31LAGIEE325 pKa = 3.5GWMAKK330 pKa = 9.98RR331 pKa = 11.84GFRR334 pKa = 11.84LVTEE338 pKa = 4.27EE339 pKa = 3.99PVYY342 pKa = 10.42EE343 pKa = 4.03INRR346 pKa = 11.84VEE348 pKa = 4.3FCQCRR353 pKa = 11.84YY354 pKa = 9.22MDD356 pKa = 4.21TVPPTMVRR364 pKa = 11.84NPLKK368 pKa = 10.77AITQDD373 pKa = 3.48HH374 pKa = 6.24AWIEE378 pKa = 4.23DD379 pKa = 3.41RR380 pKa = 11.84SISYY384 pKa = 11.09AEE386 pKa = 3.91VLAATGLGGLSLYY399 pKa = 10.74GHH401 pKa = 6.79IPVLGAYY408 pKa = 9.76YY409 pKa = 10.7DD410 pKa = 3.77LLARR414 pKa = 11.84TTQPSRR420 pKa = 11.84RR421 pKa = 11.84TLNRR425 pKa = 11.84LDD427 pKa = 5.17FRR429 pKa = 11.84SSWLRR434 pKa = 11.84DD435 pKa = 3.04ATMSGGFTEE444 pKa = 4.66PSEE447 pKa = 3.92QARR450 pKa = 11.84YY451 pKa = 9.36QFWLTWGMSPGEE463 pKa = 4.08QRR465 pKa = 11.84AHH467 pKa = 5.49EE468 pKa = 4.27MNFRR472 pKa = 11.84ACDD475 pKa = 3.76LQDD478 pKa = 4.17LVASDD483 pKa = 5.2TITKK487 pKa = 8.4ATRR490 pKa = 11.84NEE492 pKa = 3.82KK493 pKa = 9.31TDD495 pKa = 3.64PYY497 pKa = 10.47GNNYY501 pKa = 9.37YY502 pKa = 10.77LL503 pKa = 4.42
Molecular weight: 57.17 kDa
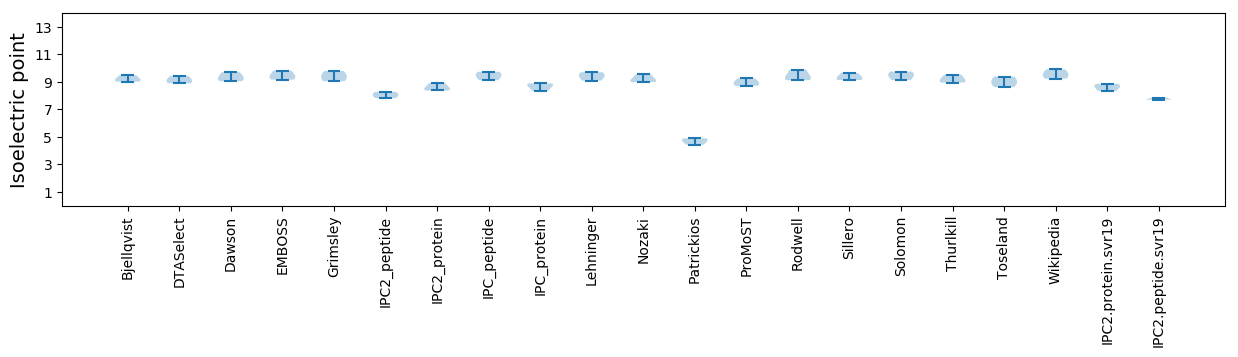
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG56|A0A1L3KG56_9VIRU Uncharacterized protein OS=Changjiang crawfish virus 7 OX=1922771 PE=4 SV=1
MM1 pKa = 7.35VKK3 pKa = 9.67TKK5 pKa = 8.52GTKK8 pKa = 10.2GKK10 pKa = 10.05GKK12 pKa = 9.0SAKK15 pKa = 9.68RR16 pKa = 11.84VAKK19 pKa = 9.95RR20 pKa = 11.84PAMRR24 pKa = 11.84VRR26 pKa = 11.84GPEE29 pKa = 3.65GHH31 pKa = 6.27EE32 pKa = 3.68ARR34 pKa = 11.84LARR37 pKa = 11.84LIMDD41 pKa = 4.32PCGAEE46 pKa = 3.99LTTGYY51 pKa = 11.49ALATEE56 pKa = 4.63GLVQRR61 pKa = 11.84FNRR64 pKa = 11.84FITPVATTEE73 pKa = 4.15TNFAYY78 pKa = 9.73IFNPLSHH85 pKa = 6.73AAEE88 pKa = 4.61SIVQKK93 pKa = 10.93LSTGTGAATNISSTAPGEE111 pKa = 4.32AYY113 pKa = 10.84LDD115 pKa = 3.92ANADD119 pKa = 3.6AVSTVAACMQILYY132 pKa = 9.14TGKK135 pKa = 9.85LVDD138 pKa = 3.44RR139 pKa = 11.84KK140 pKa = 10.71GYY142 pKa = 10.2IGVCQAPWFVMNDD155 pKa = 2.89IATGTTDD162 pKa = 4.52LPTLLSYY169 pKa = 10.63CQAIQPVGSEE179 pKa = 3.8TLEE182 pKa = 3.97IKK184 pKa = 10.72FSPTIRR190 pKa = 11.84SLLGQTANVEE200 pKa = 4.27TSGGIDD206 pKa = 3.21NVLMVVAIGVDD217 pKa = 3.44PNQFVVKK224 pKa = 8.29FTSVYY229 pKa = 9.9EE230 pKa = 4.01YY231 pKa = 11.03VPKK234 pKa = 10.42FALGAPAPRR243 pKa = 11.84ATKK246 pKa = 9.75TYY248 pKa = 10.45IPGAPEE254 pKa = 4.65RR255 pKa = 11.84IVSTLDD261 pKa = 3.37RR262 pKa = 11.84LGHH265 pKa = 5.54WWHH268 pKa = 6.35NAGNAAAAAYY278 pKa = 9.99RR279 pKa = 11.84LGGQMVYY286 pKa = 10.75GAGQAARR293 pKa = 11.84LVSATVNTARR303 pKa = 11.84TLRR306 pKa = 11.84SAAVPLLALTGG317 pKa = 3.73
MM1 pKa = 7.35VKK3 pKa = 9.67TKK5 pKa = 8.52GTKK8 pKa = 10.2GKK10 pKa = 10.05GKK12 pKa = 9.0SAKK15 pKa = 9.68RR16 pKa = 11.84VAKK19 pKa = 9.95RR20 pKa = 11.84PAMRR24 pKa = 11.84VRR26 pKa = 11.84GPEE29 pKa = 3.65GHH31 pKa = 6.27EE32 pKa = 3.68ARR34 pKa = 11.84LARR37 pKa = 11.84LIMDD41 pKa = 4.32PCGAEE46 pKa = 3.99LTTGYY51 pKa = 11.49ALATEE56 pKa = 4.63GLVQRR61 pKa = 11.84FNRR64 pKa = 11.84FITPVATTEE73 pKa = 4.15TNFAYY78 pKa = 9.73IFNPLSHH85 pKa = 6.73AAEE88 pKa = 4.61SIVQKK93 pKa = 10.93LSTGTGAATNISSTAPGEE111 pKa = 4.32AYY113 pKa = 10.84LDD115 pKa = 3.92ANADD119 pKa = 3.6AVSTVAACMQILYY132 pKa = 9.14TGKK135 pKa = 9.85LVDD138 pKa = 3.44RR139 pKa = 11.84KK140 pKa = 10.71GYY142 pKa = 10.2IGVCQAPWFVMNDD155 pKa = 2.89IATGTTDD162 pKa = 4.52LPTLLSYY169 pKa = 10.63CQAIQPVGSEE179 pKa = 3.8TLEE182 pKa = 3.97IKK184 pKa = 10.72FSPTIRR190 pKa = 11.84SLLGQTANVEE200 pKa = 4.27TSGGIDD206 pKa = 3.21NVLMVVAIGVDD217 pKa = 3.44PNQFVVKK224 pKa = 8.29FTSVYY229 pKa = 9.9EE230 pKa = 4.01YY231 pKa = 11.03VPKK234 pKa = 10.42FALGAPAPRR243 pKa = 11.84ATKK246 pKa = 9.75TYY248 pKa = 10.45IPGAPEE254 pKa = 4.65RR255 pKa = 11.84IVSTLDD261 pKa = 3.37RR262 pKa = 11.84LGHH265 pKa = 5.54WWHH268 pKa = 6.35NAGNAAAAAYY278 pKa = 9.99RR279 pKa = 11.84LGGQMVYY286 pKa = 10.75GAGQAARR293 pKa = 11.84LVSATVNTARR303 pKa = 11.84TLRR306 pKa = 11.84SAAVPLLALTGG317 pKa = 3.73
Molecular weight: 33.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1029 |
209 |
503 |
343.0 |
37.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.856 ± 2.949 | 0.972 ± 0.155 |
4.373 ± 0.744 | 5.637 ± 0.651 |
2.721 ± 0.276 | 8.941 ± 0.634 |
2.527 ± 0.708 | 3.596 ± 0.415 |
3.984 ± 0.517 | 8.552 ± 0.13 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.624 ± 0.228 | 2.721 ± 0.437 |
4.47 ± 0.376 | 3.596 ± 0.56 |
8.066 ± 0.93 | 5.539 ± 0.93 |
6.511 ± 1.146 | 7.289 ± 0.487 |
1.944 ± 0.342 | 4.082 ± 0.615 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
