
bacterium 336/3
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; unclassified Bacteroidetes/Chlorobi group
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
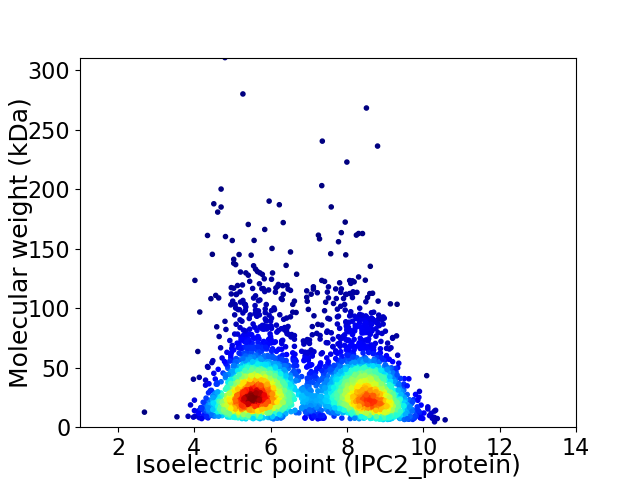
Virtual 2D-PAGE plot for 4024 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
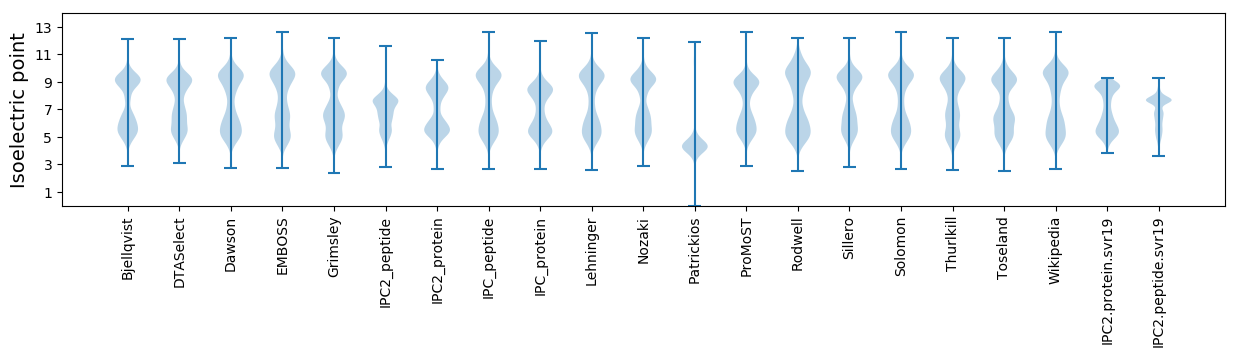
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M9DN07|A0A0M9DN07_9BACT MarR family transcriptional regulator OS=bacterium 336/3 OX=1664068 GN=AD998_00065 PE=4 SV=1
MM1 pKa = 7.42FSYY4 pKa = 10.29IGNDD8 pKa = 3.58KK9 pKa = 10.96LCLIHH14 pKa = 7.23FKK16 pKa = 10.34ILKK19 pKa = 10.1NMDD22 pKa = 4.05GIEE25 pKa = 3.93QSFEE29 pKa = 4.14EE30 pKa = 4.65VFKK33 pKa = 10.75EE34 pKa = 4.19AYY36 pKa = 8.82EE37 pKa = 3.93QEE39 pKa = 4.34EE40 pKa = 4.47FNGVVVVAHH49 pKa = 5.9QGEE52 pKa = 4.32ALYY55 pKa = 10.93EE56 pKa = 4.08EE57 pKa = 4.96AFGIADD63 pKa = 3.67IEE65 pKa = 4.48TEE67 pKa = 3.98EE68 pKa = 4.0NLAVNSIFNLASVSKK83 pKa = 10.53QFTATCIMLLYY94 pKa = 9.85EE95 pKa = 4.4QEE97 pKa = 4.15KK98 pKa = 10.75LDD100 pKa = 4.02YY101 pKa = 10.91DD102 pKa = 4.32DD103 pKa = 6.79AITTYY108 pKa = 9.4LTNLPYY114 pKa = 11.06DD115 pKa = 4.25NITIRR120 pKa = 11.84HH121 pKa = 6.21LLNHH125 pKa = 6.41TSGLPDD131 pKa = 3.36YY132 pKa = 10.49EE133 pKa = 5.56DD134 pKa = 3.01IVEE137 pKa = 4.6EE138 pKa = 4.28YY139 pKa = 10.32WDD141 pKa = 4.22GDD143 pKa = 3.77TEE145 pKa = 5.19KK146 pKa = 11.26DD147 pKa = 3.27FFTIKK152 pKa = 10.59DD153 pKa = 3.65LLEE156 pKa = 4.51IYY158 pKa = 10.42SGEE161 pKa = 4.03EE162 pKa = 3.81LEE164 pKa = 5.82LDD166 pKa = 4.18FEE168 pKa = 4.62PEE170 pKa = 3.56EE171 pKa = 4.44KK172 pKa = 10.71YY173 pKa = 10.61EE174 pKa = 3.95YY175 pKa = 11.1SNTGYY180 pKa = 10.97VFLACIVEE188 pKa = 4.45KK189 pKa = 10.79ASGMSFEE196 pKa = 5.5DD197 pKa = 4.65FIKK200 pKa = 10.54KK201 pKa = 10.03YY202 pKa = 10.25VFEE205 pKa = 4.27PLGMKK210 pKa = 10.43NSFACRR216 pKa = 11.84KK217 pKa = 8.91PNKK220 pKa = 8.4TPKK223 pKa = 10.02KK224 pKa = 8.45AAQGFTTNDD233 pKa = 3.19DD234 pKa = 3.48GDD236 pKa = 4.44YY237 pKa = 11.44EE238 pKa = 5.03DD239 pKa = 3.6YY240 pKa = 10.62TYY242 pKa = 11.35NYY244 pKa = 10.58LDD246 pKa = 4.0GLVGDD251 pKa = 4.69GNIFSSVSDD260 pKa = 3.36LVIYY264 pKa = 10.23AHH266 pKa = 7.07ALINGDD272 pKa = 4.96LLSQDD277 pKa = 3.86TLDD280 pKa = 3.94EE281 pKa = 4.61AFTPATLANGKK292 pKa = 8.35KK293 pKa = 5.41TTYY296 pKa = 10.45GFGWEE301 pKa = 4.19LEE303 pKa = 4.22GEE305 pKa = 4.51GFISHH310 pKa = 6.57EE311 pKa = 4.46GSWEE315 pKa = 4.24GYY317 pKa = 7.36NTYY320 pKa = 10.77LGIDD324 pKa = 3.78LEE326 pKa = 4.62DD327 pKa = 4.69GYY329 pKa = 11.92VFVVLDD335 pKa = 4.3NGDD338 pKa = 3.91HH339 pKa = 7.25PDD341 pKa = 3.32IADD344 pKa = 3.75LVEE347 pKa = 3.87EE348 pKa = 4.22AMEE351 pKa = 4.41GFYY354 pKa = 10.59EE355 pKa = 4.22
MM1 pKa = 7.42FSYY4 pKa = 10.29IGNDD8 pKa = 3.58KK9 pKa = 10.96LCLIHH14 pKa = 7.23FKK16 pKa = 10.34ILKK19 pKa = 10.1NMDD22 pKa = 4.05GIEE25 pKa = 3.93QSFEE29 pKa = 4.14EE30 pKa = 4.65VFKK33 pKa = 10.75EE34 pKa = 4.19AYY36 pKa = 8.82EE37 pKa = 3.93QEE39 pKa = 4.34EE40 pKa = 4.47FNGVVVVAHH49 pKa = 5.9QGEE52 pKa = 4.32ALYY55 pKa = 10.93EE56 pKa = 4.08EE57 pKa = 4.96AFGIADD63 pKa = 3.67IEE65 pKa = 4.48TEE67 pKa = 3.98EE68 pKa = 4.0NLAVNSIFNLASVSKK83 pKa = 10.53QFTATCIMLLYY94 pKa = 9.85EE95 pKa = 4.4QEE97 pKa = 4.15KK98 pKa = 10.75LDD100 pKa = 4.02YY101 pKa = 10.91DD102 pKa = 4.32DD103 pKa = 6.79AITTYY108 pKa = 9.4LTNLPYY114 pKa = 11.06DD115 pKa = 4.25NITIRR120 pKa = 11.84HH121 pKa = 6.21LLNHH125 pKa = 6.41TSGLPDD131 pKa = 3.36YY132 pKa = 10.49EE133 pKa = 5.56DD134 pKa = 3.01IVEE137 pKa = 4.6EE138 pKa = 4.28YY139 pKa = 10.32WDD141 pKa = 4.22GDD143 pKa = 3.77TEE145 pKa = 5.19KK146 pKa = 11.26DD147 pKa = 3.27FFTIKK152 pKa = 10.59DD153 pKa = 3.65LLEE156 pKa = 4.51IYY158 pKa = 10.42SGEE161 pKa = 4.03EE162 pKa = 3.81LEE164 pKa = 5.82LDD166 pKa = 4.18FEE168 pKa = 4.62PEE170 pKa = 3.56EE171 pKa = 4.44KK172 pKa = 10.71YY173 pKa = 10.61EE174 pKa = 3.95YY175 pKa = 11.1SNTGYY180 pKa = 10.97VFLACIVEE188 pKa = 4.45KK189 pKa = 10.79ASGMSFEE196 pKa = 5.5DD197 pKa = 4.65FIKK200 pKa = 10.54KK201 pKa = 10.03YY202 pKa = 10.25VFEE205 pKa = 4.27PLGMKK210 pKa = 10.43NSFACRR216 pKa = 11.84KK217 pKa = 8.91PNKK220 pKa = 8.4TPKK223 pKa = 10.02KK224 pKa = 8.45AAQGFTTNDD233 pKa = 3.19DD234 pKa = 3.48GDD236 pKa = 4.44YY237 pKa = 11.44EE238 pKa = 5.03DD239 pKa = 3.6YY240 pKa = 10.62TYY242 pKa = 11.35NYY244 pKa = 10.58LDD246 pKa = 4.0GLVGDD251 pKa = 4.69GNIFSSVSDD260 pKa = 3.36LVIYY264 pKa = 10.23AHH266 pKa = 7.07ALINGDD272 pKa = 4.96LLSQDD277 pKa = 3.86TLDD280 pKa = 3.94EE281 pKa = 4.61AFTPATLANGKK292 pKa = 8.35KK293 pKa = 5.41TTYY296 pKa = 10.45GFGWEE301 pKa = 4.19LEE303 pKa = 4.22GEE305 pKa = 4.51GFISHH310 pKa = 6.57EE311 pKa = 4.46GSWEE315 pKa = 4.24GYY317 pKa = 7.36NTYY320 pKa = 10.77LGIDD324 pKa = 3.78LEE326 pKa = 4.62DD327 pKa = 4.69GYY329 pKa = 11.92VFVVLDD335 pKa = 4.3NGDD338 pKa = 3.91HH339 pKa = 7.25PDD341 pKa = 3.32IADD344 pKa = 3.75LVEE347 pKa = 3.87EE348 pKa = 4.22AMEE351 pKa = 4.41GFYY354 pKa = 10.59EE355 pKa = 4.22
Molecular weight: 40.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0N0UYQ3|A0A0N0UYQ3_9BACT Uncharacterized protein OS=bacterium 336/3 OX=1664068 GN=AD998_16430 PE=4 SV=1
MM1 pKa = 7.04KK2 pKa = 9.96TVFLTFLVLITLPAFAQEE20 pKa = 4.73GEE22 pKa = 4.18ILHH25 pKa = 7.27DD26 pKa = 3.8GLFWGIAGTNITKK39 pKa = 10.37QNSPWGVDD47 pKa = 3.1LSVQNRR53 pKa = 11.84MYY55 pKa = 11.25KK56 pKa = 10.17NFSRR60 pKa = 11.84LQTILGTVHH69 pKa = 6.94LNYY72 pKa = 9.93RR73 pKa = 11.84RR74 pKa = 11.84PNQPWGFAVGYY85 pKa = 9.16IGGVFEE91 pKa = 4.69PLGVLHH97 pKa = 7.15IAQGRR102 pKa = 11.84INRR105 pKa = 11.84FGTLANGKK113 pKa = 9.05IPFVARR119 pKa = 11.84LSYY122 pKa = 10.92DD123 pKa = 3.7RR124 pKa = 11.84LWANPIYY131 pKa = 9.07EE132 pKa = 4.4TDD134 pKa = 2.83VRR136 pKa = 11.84TPVNHH141 pKa = 6.79RR142 pKa = 11.84WRR144 pKa = 11.84LLGQITPPITKK155 pKa = 9.92KK156 pKa = 10.53ISGFLAFEE164 pKa = 3.91PFIIRR169 pKa = 11.84RR170 pKa = 11.84GLWFSEE176 pKa = 3.91TRR178 pKa = 11.84GQAGFRR184 pKa = 11.84FLMKK188 pKa = 10.38NANLDD193 pKa = 3.27VLYY196 pKa = 10.43FNRR199 pKa = 11.84WLKK202 pKa = 10.32PINEE206 pKa = 4.25RR207 pKa = 11.84DD208 pKa = 3.3LTRR211 pKa = 11.84FEE213 pKa = 5.43HH214 pKa = 5.79VFQVIYY220 pKa = 7.44MHH222 pKa = 7.62RR223 pKa = 11.84IQFKK227 pKa = 10.29SNKK230 pKa = 5.73QTEE233 pKa = 4.14NSTNN237 pKa = 3.25
MM1 pKa = 7.04KK2 pKa = 9.96TVFLTFLVLITLPAFAQEE20 pKa = 4.73GEE22 pKa = 4.18ILHH25 pKa = 7.27DD26 pKa = 3.8GLFWGIAGTNITKK39 pKa = 10.37QNSPWGVDD47 pKa = 3.1LSVQNRR53 pKa = 11.84MYY55 pKa = 11.25KK56 pKa = 10.17NFSRR60 pKa = 11.84LQTILGTVHH69 pKa = 6.94LNYY72 pKa = 9.93RR73 pKa = 11.84RR74 pKa = 11.84PNQPWGFAVGYY85 pKa = 9.16IGGVFEE91 pKa = 4.69PLGVLHH97 pKa = 7.15IAQGRR102 pKa = 11.84INRR105 pKa = 11.84FGTLANGKK113 pKa = 9.05IPFVARR119 pKa = 11.84LSYY122 pKa = 10.92DD123 pKa = 3.7RR124 pKa = 11.84LWANPIYY131 pKa = 9.07EE132 pKa = 4.4TDD134 pKa = 2.83VRR136 pKa = 11.84TPVNHH141 pKa = 6.79RR142 pKa = 11.84WRR144 pKa = 11.84LLGQITPPITKK155 pKa = 9.92KK156 pKa = 10.53ISGFLAFEE164 pKa = 3.91PFIIRR169 pKa = 11.84RR170 pKa = 11.84GLWFSEE176 pKa = 3.91TRR178 pKa = 11.84GQAGFRR184 pKa = 11.84FLMKK188 pKa = 10.38NANLDD193 pKa = 3.27VLYY196 pKa = 10.43FNRR199 pKa = 11.84WLKK202 pKa = 10.32PINEE206 pKa = 4.25RR207 pKa = 11.84DD208 pKa = 3.3LTRR211 pKa = 11.84FEE213 pKa = 5.43HH214 pKa = 5.79VFQVIYY220 pKa = 7.44MHH222 pKa = 7.62RR223 pKa = 11.84IQFKK227 pKa = 10.29SNKK230 pKa = 5.73QTEE233 pKa = 4.14NSTNN237 pKa = 3.25
Molecular weight: 27.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1296309 |
38 |
2933 |
322.1 |
36.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.023 ± 0.037 | 0.828 ± 0.012 |
4.915 ± 0.028 | 6.806 ± 0.047 |
5.401 ± 0.035 | 5.906 ± 0.042 |
1.767 ± 0.018 | 8.265 ± 0.037 |
8.531 ± 0.053 | 9.549 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.196 ± 0.019 | 5.978 ± 0.038 |
3.294 ± 0.025 | 4.184 ± 0.025 |
3.277 ± 0.023 | 6.224 ± 0.031 |
5.599 ± 0.047 | 5.799 ± 0.033 |
1.162 ± 0.016 | 4.293 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
