
Flavobacterium arcticum
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
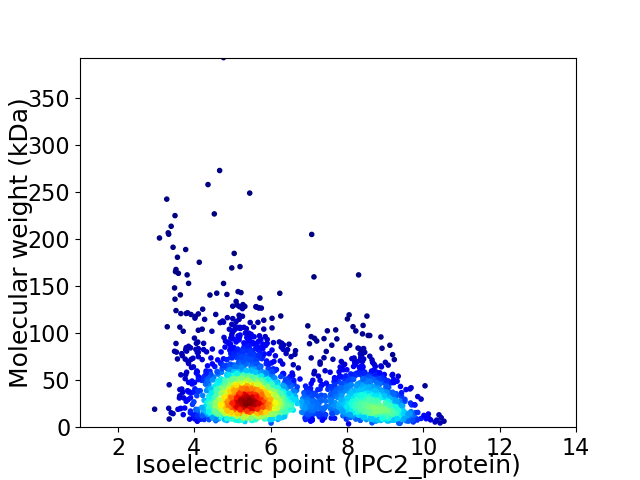
Virtual 2D-PAGE plot for 2621 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345H9I7|A0A345H9I7_9FLAO FAD-binding oxidoreductase OS=Flavobacterium arcticum OX=1784713 GN=DVK85_02990 PE=4 SV=1
MM1 pKa = 7.63KK2 pKa = 10.32KK3 pKa = 9.85QLLFLLLLITTAASAQIVNVPDD25 pKa = 3.51VAFKK29 pKa = 10.57EE30 pKa = 4.32ALLSNYY36 pKa = 9.13TDD38 pKa = 3.98AKK40 pKa = 10.6DD41 pKa = 3.48LNGNIISLDD50 pKa = 3.57ANNDD54 pKa = 3.46GEE56 pKa = 4.49IQLSEE61 pKa = 4.06ALNVGEE67 pKa = 4.09ITIDD71 pKa = 3.3LFSFFVFSLEE81 pKa = 4.64GIGSFSNLTKK91 pKa = 10.93LSITEE96 pKa = 4.03ADD98 pKa = 3.33QLLDD102 pKa = 4.03LDD104 pKa = 4.46LTALTQLTQLSISDD118 pKa = 3.9SNEE121 pKa = 3.21FDD123 pKa = 4.13SINLNGLSNLEE134 pKa = 4.17NINFNGLTGGGDD146 pKa = 3.3SMLVLNVSGVNNLTHH161 pKa = 6.89VNFTDD166 pKa = 3.61VVFDD170 pKa = 3.8MDD172 pKa = 4.96FSQLPSLEE180 pKa = 4.14SLILNNTSVYY190 pKa = 10.66FDD192 pKa = 4.3LSANNNLSYY201 pKa = 11.42VSITNPQYY209 pKa = 11.34VDD211 pKa = 3.43YY212 pKa = 10.31FLLGNKK218 pKa = 9.22PLLTYY223 pKa = 11.12LNLNFNGGIGIVNDD237 pKa = 3.88IDD239 pKa = 5.57LSGCTALDD247 pKa = 3.63DD248 pKa = 4.12LFLRR252 pKa = 11.84FDD254 pKa = 3.65EE255 pKa = 4.84AVAIIPIPFRR265 pKa = 11.84TLNLKK270 pKa = 10.51NGISNYY276 pKa = 10.7DD277 pKa = 3.36SFTVMLNNGNLQPIYY292 pKa = 10.86VCIDD296 pKa = 3.37EE297 pKa = 5.01GNEE300 pKa = 4.06TLFSEE305 pKa = 5.22TVLNDD310 pKa = 3.59PNVLISSYY318 pKa = 11.6CNFTPGGDD326 pKa = 3.68YY327 pKa = 10.18NTITGNFTFDD337 pKa = 4.45ADD339 pKa = 5.19NDD341 pKa = 4.17GCDD344 pKa = 4.44INDD347 pKa = 3.4NLINHH352 pKa = 5.97TRR354 pKa = 11.84VDD356 pKa = 3.23ITTEE360 pKa = 3.7NDD362 pKa = 2.85EE363 pKa = 4.22EE364 pKa = 4.29FAVFSNNIGEE374 pKa = 4.38YY375 pKa = 9.62NFYY378 pKa = 10.11WGAGTFTLSPSIEE391 pKa = 4.09NNDD394 pKa = 3.27WFTITPSSATVTFEE408 pKa = 5.03DD409 pKa = 4.42DD410 pKa = 3.45NNNIEE415 pKa = 4.3TQSFCVTPNGEE426 pKa = 4.19HH427 pKa = 7.21PDD429 pKa = 3.62VEE431 pKa = 4.76VVIVPTEE438 pKa = 3.67PAQPGFNASYY448 pKa = 10.62KK449 pKa = 8.27ITYY452 pKa = 9.1RR453 pKa = 11.84NKK455 pKa = 10.29GNQTLSGNVLLTYY468 pKa = 10.76DD469 pKa = 4.22EE470 pKa = 4.65TVLDD474 pKa = 4.36YY475 pKa = 11.37VSSTSGVGAATNTFGWSYY493 pKa = 11.85SDD495 pKa = 3.55LQPFEE500 pKa = 4.17TRR502 pKa = 11.84NINLTLNVNAPTDD515 pKa = 3.87TPAVNIDD522 pKa = 3.8DD523 pKa = 5.4LLMFTANITSATGDD537 pKa = 4.01EE538 pKa = 4.47NPDD541 pKa = 3.73DD542 pKa = 3.94NTFTLAQTVVGSFDD556 pKa = 4.41PNDD559 pKa = 3.42ITCLEE564 pKa = 4.44GEE566 pKa = 4.36AVHH569 pKa = 6.16VSKK572 pKa = 10.9IGDD575 pKa = 3.58YY576 pKa = 10.9LHH578 pKa = 6.9YY579 pKa = 10.74NINFEE584 pKa = 4.42NTGTAPATFIVVKK597 pKa = 10.91NEE599 pKa = 3.49INEE602 pKa = 4.13EE603 pKa = 4.0QFDD606 pKa = 4.04VSTLQLIDD614 pKa = 3.7ASHH617 pKa = 5.82NVEE620 pKa = 4.27TRR622 pKa = 11.84ITGNKK627 pKa = 8.53VEE629 pKa = 5.19FYY631 pKa = 10.7FGNINLAANGGKK643 pKa = 10.65GNVLYY648 pKa = 10.43KK649 pKa = 10.47IKK651 pKa = 9.7TLNTLQEE658 pKa = 4.58DD659 pKa = 4.72DD660 pKa = 4.84IVMQEE665 pKa = 3.5ANIYY669 pKa = 10.21FDD671 pKa = 3.94YY672 pKa = 11.07NFPIEE677 pKa = 4.2TNEE680 pKa = 3.81ANTTFAVLSNDD691 pKa = 3.26VLTKK695 pKa = 10.58DD696 pKa = 4.86DD697 pKa = 4.28SVKK700 pKa = 10.61LYY702 pKa = 9.79PNPTTDD708 pKa = 3.26NVTIAANSAITSVQLYY724 pKa = 9.78DD725 pKa = 3.45VQGRR729 pKa = 11.84LLHH732 pKa = 6.23SRR734 pKa = 11.84VGNEE738 pKa = 3.34TSIRR742 pKa = 11.84LDD744 pKa = 3.22VSAQPSGLYY753 pKa = 9.34FVKK756 pKa = 9.5ITTDD760 pKa = 2.98NGSKK764 pKa = 10.19VEE766 pKa = 4.07KK767 pKa = 10.2LVKK770 pKa = 9.94EE771 pKa = 4.23
MM1 pKa = 7.63KK2 pKa = 10.32KK3 pKa = 9.85QLLFLLLLITTAASAQIVNVPDD25 pKa = 3.51VAFKK29 pKa = 10.57EE30 pKa = 4.32ALLSNYY36 pKa = 9.13TDD38 pKa = 3.98AKK40 pKa = 10.6DD41 pKa = 3.48LNGNIISLDD50 pKa = 3.57ANNDD54 pKa = 3.46GEE56 pKa = 4.49IQLSEE61 pKa = 4.06ALNVGEE67 pKa = 4.09ITIDD71 pKa = 3.3LFSFFVFSLEE81 pKa = 4.64GIGSFSNLTKK91 pKa = 10.93LSITEE96 pKa = 4.03ADD98 pKa = 3.33QLLDD102 pKa = 4.03LDD104 pKa = 4.46LTALTQLTQLSISDD118 pKa = 3.9SNEE121 pKa = 3.21FDD123 pKa = 4.13SINLNGLSNLEE134 pKa = 4.17NINFNGLTGGGDD146 pKa = 3.3SMLVLNVSGVNNLTHH161 pKa = 6.89VNFTDD166 pKa = 3.61VVFDD170 pKa = 3.8MDD172 pKa = 4.96FSQLPSLEE180 pKa = 4.14SLILNNTSVYY190 pKa = 10.66FDD192 pKa = 4.3LSANNNLSYY201 pKa = 11.42VSITNPQYY209 pKa = 11.34VDD211 pKa = 3.43YY212 pKa = 10.31FLLGNKK218 pKa = 9.22PLLTYY223 pKa = 11.12LNLNFNGGIGIVNDD237 pKa = 3.88IDD239 pKa = 5.57LSGCTALDD247 pKa = 3.63DD248 pKa = 4.12LFLRR252 pKa = 11.84FDD254 pKa = 3.65EE255 pKa = 4.84AVAIIPIPFRR265 pKa = 11.84TLNLKK270 pKa = 10.51NGISNYY276 pKa = 10.7DD277 pKa = 3.36SFTVMLNNGNLQPIYY292 pKa = 10.86VCIDD296 pKa = 3.37EE297 pKa = 5.01GNEE300 pKa = 4.06TLFSEE305 pKa = 5.22TVLNDD310 pKa = 3.59PNVLISSYY318 pKa = 11.6CNFTPGGDD326 pKa = 3.68YY327 pKa = 10.18NTITGNFTFDD337 pKa = 4.45ADD339 pKa = 5.19NDD341 pKa = 4.17GCDD344 pKa = 4.44INDD347 pKa = 3.4NLINHH352 pKa = 5.97TRR354 pKa = 11.84VDD356 pKa = 3.23ITTEE360 pKa = 3.7NDD362 pKa = 2.85EE363 pKa = 4.22EE364 pKa = 4.29FAVFSNNIGEE374 pKa = 4.38YY375 pKa = 9.62NFYY378 pKa = 10.11WGAGTFTLSPSIEE391 pKa = 4.09NNDD394 pKa = 3.27WFTITPSSATVTFEE408 pKa = 5.03DD409 pKa = 4.42DD410 pKa = 3.45NNNIEE415 pKa = 4.3TQSFCVTPNGEE426 pKa = 4.19HH427 pKa = 7.21PDD429 pKa = 3.62VEE431 pKa = 4.76VVIVPTEE438 pKa = 3.67PAQPGFNASYY448 pKa = 10.62KK449 pKa = 8.27ITYY452 pKa = 9.1RR453 pKa = 11.84NKK455 pKa = 10.29GNQTLSGNVLLTYY468 pKa = 10.76DD469 pKa = 4.22EE470 pKa = 4.65TVLDD474 pKa = 4.36YY475 pKa = 11.37VSSTSGVGAATNTFGWSYY493 pKa = 11.85SDD495 pKa = 3.55LQPFEE500 pKa = 4.17TRR502 pKa = 11.84NINLTLNVNAPTDD515 pKa = 3.87TPAVNIDD522 pKa = 3.8DD523 pKa = 5.4LLMFTANITSATGDD537 pKa = 4.01EE538 pKa = 4.47NPDD541 pKa = 3.73DD542 pKa = 3.94NTFTLAQTVVGSFDD556 pKa = 4.41PNDD559 pKa = 3.42ITCLEE564 pKa = 4.44GEE566 pKa = 4.36AVHH569 pKa = 6.16VSKK572 pKa = 10.9IGDD575 pKa = 3.58YY576 pKa = 10.9LHH578 pKa = 6.9YY579 pKa = 10.74NINFEE584 pKa = 4.42NTGTAPATFIVVKK597 pKa = 10.91NEE599 pKa = 3.49INEE602 pKa = 4.13EE603 pKa = 4.0QFDD606 pKa = 4.04VSTLQLIDD614 pKa = 3.7ASHH617 pKa = 5.82NVEE620 pKa = 4.27TRR622 pKa = 11.84ITGNKK627 pKa = 8.53VEE629 pKa = 5.19FYY631 pKa = 10.7FGNINLAANGGKK643 pKa = 10.65GNVLYY648 pKa = 10.43KK649 pKa = 10.47IKK651 pKa = 9.7TLNTLQEE658 pKa = 4.58DD659 pKa = 4.72DD660 pKa = 4.84IVMQEE665 pKa = 3.5ANIYY669 pKa = 10.21FDD671 pKa = 3.94YY672 pKa = 11.07NFPIEE677 pKa = 4.2TNEE680 pKa = 3.81ANTTFAVLSNDD691 pKa = 3.26VLTKK695 pKa = 10.58DD696 pKa = 4.86DD697 pKa = 4.28SVKK700 pKa = 10.61LYY702 pKa = 9.79PNPTTDD708 pKa = 3.26NVTIAANSAITSVQLYY724 pKa = 9.78DD725 pKa = 3.45VQGRR729 pKa = 11.84LLHH732 pKa = 6.23SRR734 pKa = 11.84VGNEE738 pKa = 3.34TSIRR742 pKa = 11.84LDD744 pKa = 3.22VSAQPSGLYY753 pKa = 9.34FVKK756 pKa = 9.5ITTDD760 pKa = 2.98NGSKK764 pKa = 10.19VEE766 pKa = 4.07KK767 pKa = 10.2LVKK770 pKa = 9.94EE771 pKa = 4.23
Molecular weight: 84.54 kDa
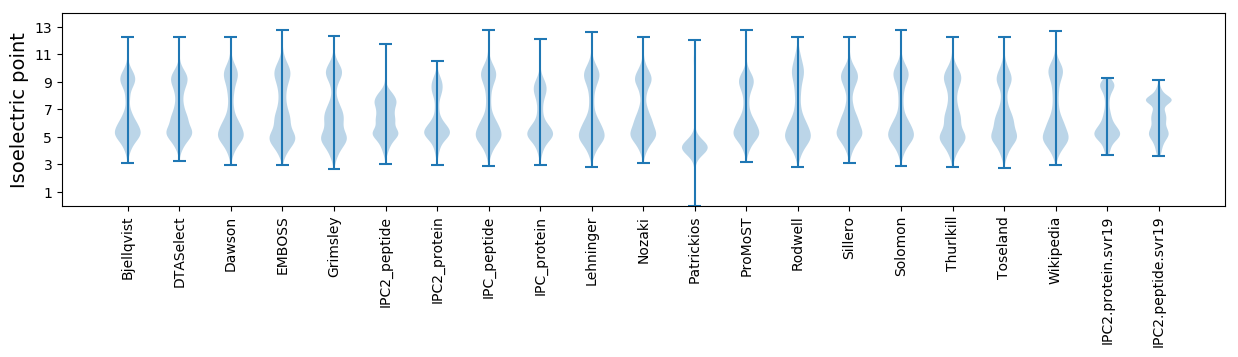
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345H9F1|A0A345H9F1_9FLAO 50S ribosomal protein L23 OS=Flavobacterium arcticum OX=1784713 GN=rplW PE=3 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNSFDD22 pKa = 3.97TITTDD27 pKa = 2.9KK28 pKa = 10.84PEE30 pKa = 4.41RR31 pKa = 11.84SLIAPKK37 pKa = 10.39KK38 pKa = 9.8NSGGRR43 pKa = 11.84NSQGKK48 pKa = 6.33MTMRR52 pKa = 11.84YY53 pKa = 7.39TGGGHH58 pKa = 4.86KK59 pKa = 9.44QRR61 pKa = 11.84YY62 pKa = 8.74RR63 pKa = 11.84IIDD66 pKa = 3.82FKK68 pKa = 9.82RR69 pKa = 11.84TKK71 pKa = 10.35VGIPAVVKK79 pKa = 9.88TIEE82 pKa = 3.93YY83 pKa = 10.33DD84 pKa = 3.47PNRR87 pKa = 11.84SAFISLLYY95 pKa = 10.27YY96 pKa = 10.82ADD98 pKa = 3.79GEE100 pKa = 4.22KK101 pKa = 10.52AYY103 pKa = 9.78IVAQNGLQVGQTVVSGEE120 pKa = 4.08DD121 pKa = 3.29AVPEE125 pKa = 4.25IGNAMPLSKK134 pKa = 10.36IPLGTVISCIEE145 pKa = 3.9LRR147 pKa = 11.84PGQGAVIARR156 pKa = 11.84SAGTFAQLMARR167 pKa = 11.84DD168 pKa = 4.06GKK170 pKa = 10.75YY171 pKa = 9.05ATIKK175 pKa = 9.6MPSGEE180 pKa = 4.18TRR182 pKa = 11.84LILLTCMATIGAVSNSDD199 pKa = 3.15HH200 pKa = 6.2QLVVSGKK207 pKa = 9.63AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVAMNPVDD229 pKa = 3.63HH230 pKa = 7.13PMGGGEE236 pKa = 4.08GRR238 pKa = 11.84SSGGHH243 pKa = 4.81PRR245 pKa = 11.84SRR247 pKa = 11.84KK248 pKa = 8.85GLPAKK253 pKa = 10.34GYY255 pKa = 6.98RR256 pKa = 11.84TRR258 pKa = 11.84SKK260 pKa = 10.97VNPSNKK266 pKa = 9.96YY267 pKa = 8.54IVEE270 pKa = 4.07RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.24KK274 pKa = 10.1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNSFDD22 pKa = 3.97TITTDD27 pKa = 2.9KK28 pKa = 10.84PEE30 pKa = 4.41RR31 pKa = 11.84SLIAPKK37 pKa = 10.39KK38 pKa = 9.8NSGGRR43 pKa = 11.84NSQGKK48 pKa = 6.33MTMRR52 pKa = 11.84YY53 pKa = 7.39TGGGHH58 pKa = 4.86KK59 pKa = 9.44QRR61 pKa = 11.84YY62 pKa = 8.74RR63 pKa = 11.84IIDD66 pKa = 3.82FKK68 pKa = 9.82RR69 pKa = 11.84TKK71 pKa = 10.35VGIPAVVKK79 pKa = 9.88TIEE82 pKa = 3.93YY83 pKa = 10.33DD84 pKa = 3.47PNRR87 pKa = 11.84SAFISLLYY95 pKa = 10.27YY96 pKa = 10.82ADD98 pKa = 3.79GEE100 pKa = 4.22KK101 pKa = 10.52AYY103 pKa = 9.78IVAQNGLQVGQTVVSGEE120 pKa = 4.08DD121 pKa = 3.29AVPEE125 pKa = 4.25IGNAMPLSKK134 pKa = 10.36IPLGTVISCIEE145 pKa = 3.9LRR147 pKa = 11.84PGQGAVIARR156 pKa = 11.84SAGTFAQLMARR167 pKa = 11.84DD168 pKa = 4.06GKK170 pKa = 10.75YY171 pKa = 9.05ATIKK175 pKa = 9.6MPSGEE180 pKa = 4.18TRR182 pKa = 11.84LILLTCMATIGAVSNSDD199 pKa = 3.15HH200 pKa = 6.2QLVVSGKK207 pKa = 9.63AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVAMNPVDD229 pKa = 3.63HH230 pKa = 7.13PMGGGEE236 pKa = 4.08GRR238 pKa = 11.84SSGGHH243 pKa = 4.81PRR245 pKa = 11.84SRR247 pKa = 11.84KK248 pKa = 8.85GLPAKK253 pKa = 10.34GYY255 pKa = 6.98RR256 pKa = 11.84TRR258 pKa = 11.84SKK260 pKa = 10.97VNPSNKK266 pKa = 9.96YY267 pKa = 8.54IVEE270 pKa = 4.07RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.24KK274 pKa = 10.1
Molecular weight: 29.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
896263 |
30 |
3500 |
342.0 |
38.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.752 ± 0.058 | 0.762 ± 0.015 |
5.738 ± 0.041 | 6.68 ± 0.05 |
4.947 ± 0.034 | 6.444 ± 0.044 |
1.676 ± 0.024 | 7.883 ± 0.052 |
7.282 ± 0.072 | 8.782 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.34 ± 0.025 | 6.2 ± 0.046 |
3.457 ± 0.031 | 3.319 ± 0.023 |
3.317 ± 0.036 | 6.162 ± 0.042 |
6.419 ± 0.07 | 6.458 ± 0.037 |
1.002 ± 0.017 | 4.381 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
