
Blastococcus sp. DSM 46786
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Blastococcus; unclassified Blastococcus
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
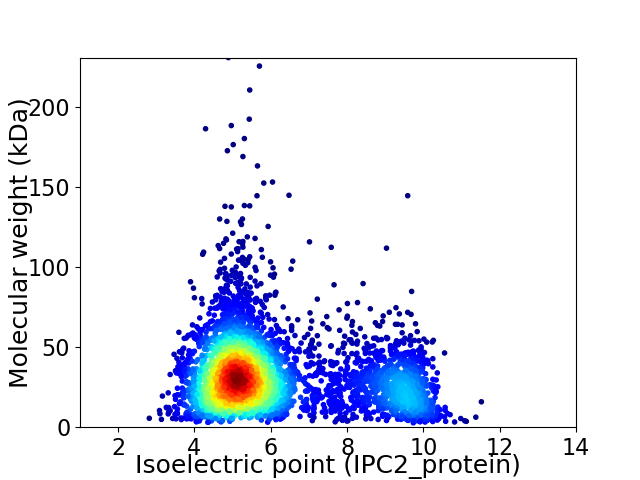
Virtual 2D-PAGE plot for 4401 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
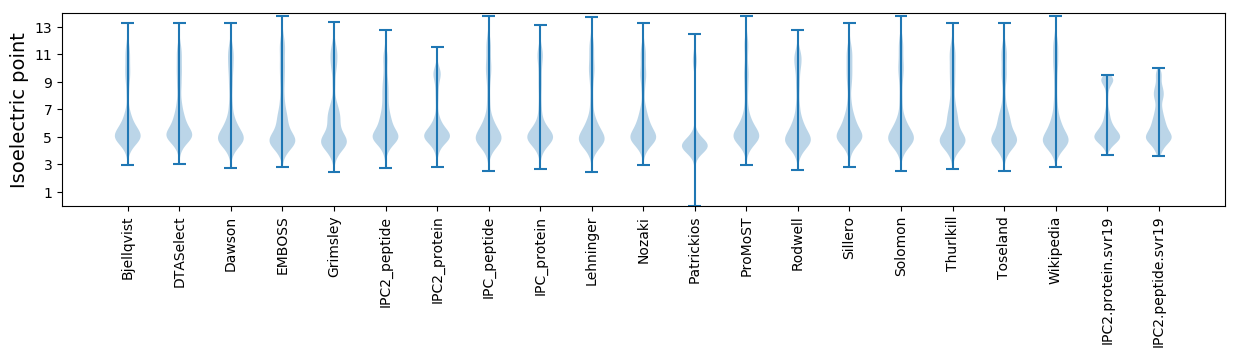
Protein with the lowest isoelectric point:
>tr|A0A1H7T451|A0A1H7T451_9ACTN Cystathionine gamma-lyase OS=Blastococcus sp. DSM 46786 OX=1798227 GN=SAMN04515665_11980 PE=3 SV=1
MM1 pKa = 7.46RR2 pKa = 11.84QLLHH6 pKa = 6.66RR7 pKa = 11.84VAPVVVAALVLTGCATGTVRR27 pKa = 11.84GQATPPGDD35 pKa = 3.63GPTDD39 pKa = 3.39VTAGEE44 pKa = 4.41FPVTGAGDD52 pKa = 3.59EE53 pKa = 5.24PIDD56 pKa = 3.64QFARR60 pKa = 11.84NALADD65 pKa = 4.7LDD67 pKa = 4.87AFWAQAYY74 pKa = 9.12PEE76 pKa = 4.39YY77 pKa = 10.4FGEE80 pKa = 4.69EE81 pKa = 4.15YY82 pKa = 10.46TPLAGGYY89 pKa = 9.97FSVDD93 pKa = 2.97SEE95 pKa = 4.6ALDD98 pKa = 3.54EE99 pKa = 4.27SAYY102 pKa = 9.67PGTGIGCEE110 pKa = 4.12GSYY113 pKa = 8.76TAPEE117 pKa = 4.24DD118 pKa = 3.55VAGNAFYY125 pKa = 11.11DD126 pKa = 4.06PVCDD130 pKa = 4.25VIAYY134 pKa = 9.42DD135 pKa = 3.9RR136 pKa = 11.84ALLQEE141 pKa = 4.59LSDD144 pKa = 3.93DD145 pKa = 3.72YY146 pKa = 11.89GRR148 pKa = 11.84FLVPVVMAHH157 pKa = 6.08EE158 pKa = 5.42FGHH161 pKa = 6.59AMQGRR166 pKa = 11.84FGFAEE171 pKa = 4.33SGRR174 pKa = 11.84SIQDD178 pKa = 2.99EE179 pKa = 4.39TQADD183 pKa = 4.26CLAGAWTRR191 pKa = 11.84WVADD195 pKa = 3.23GRR197 pKa = 11.84AAHH200 pKa = 6.64LSLRR204 pKa = 11.84EE205 pKa = 4.06PEE207 pKa = 4.32LDD209 pKa = 3.43DD210 pKa = 3.65VVRR213 pKa = 11.84GFLLLRR219 pKa = 11.84DD220 pKa = 4.48DD221 pKa = 4.65VGSDD225 pKa = 3.48PDD227 pKa = 3.9DD228 pKa = 4.26VEE230 pKa = 4.62AHH232 pKa = 5.93GSYY235 pKa = 10.53FDD237 pKa = 4.04RR238 pKa = 11.84VSAFYY243 pKa = 10.98DD244 pKa = 3.47GFEE247 pKa = 4.57RR248 pKa = 11.84GLGTCRR254 pKa = 11.84DD255 pKa = 3.59AFGEE259 pKa = 4.09DD260 pKa = 3.27RR261 pKa = 11.84LFTAAAFTPTDD272 pKa = 4.19LDD274 pKa = 3.3QGNADD279 pKa = 3.81FADD282 pKa = 3.4IVDD285 pKa = 3.93WVGATLPVFWGEE297 pKa = 3.94VFPAAFGGEE306 pKa = 4.17FEE308 pKa = 4.49EE309 pKa = 4.83PEE311 pKa = 3.65IAGFVGSAPGCDD323 pKa = 3.5GLDD326 pKa = 3.63GRR328 pKa = 11.84DD329 pKa = 3.64LGYY332 pKa = 10.72CADD335 pKa = 4.47DD336 pKa = 3.26RR337 pKa = 11.84TVYY340 pKa = 10.28IDD342 pKa = 3.58EE343 pKa = 4.59TDD345 pKa = 3.84LAVPAYY351 pKa = 10.53DD352 pKa = 4.39EE353 pKa = 4.8IGDD356 pKa = 3.96FALATAMALPYY367 pKa = 10.55ALAVRR372 pKa = 11.84DD373 pKa = 3.9QAGLPVDD380 pKa = 4.78DD381 pKa = 5.58GEE383 pKa = 4.44ATRR386 pKa = 11.84SAVCLTGWYY395 pKa = 8.42QAQWYY400 pKa = 8.51NAEE403 pKa = 3.99FSDD406 pKa = 4.64VVGAEE411 pKa = 4.07ISPGDD416 pKa = 4.05LDD418 pKa = 3.88EE419 pKa = 5.52AVQFLLRR426 pKa = 11.84YY427 pKa = 9.06GVEE430 pKa = 4.21DD431 pKa = 3.69EE432 pKa = 4.81VFPGTDD438 pKa = 2.71ASGFEE443 pKa = 4.14LVGAFRR449 pKa = 11.84NGFLDD454 pKa = 3.98GGAACGIGVV463 pKa = 3.7
MM1 pKa = 7.46RR2 pKa = 11.84QLLHH6 pKa = 6.66RR7 pKa = 11.84VAPVVVAALVLTGCATGTVRR27 pKa = 11.84GQATPPGDD35 pKa = 3.63GPTDD39 pKa = 3.39VTAGEE44 pKa = 4.41FPVTGAGDD52 pKa = 3.59EE53 pKa = 5.24PIDD56 pKa = 3.64QFARR60 pKa = 11.84NALADD65 pKa = 4.7LDD67 pKa = 4.87AFWAQAYY74 pKa = 9.12PEE76 pKa = 4.39YY77 pKa = 10.4FGEE80 pKa = 4.69EE81 pKa = 4.15YY82 pKa = 10.46TPLAGGYY89 pKa = 9.97FSVDD93 pKa = 2.97SEE95 pKa = 4.6ALDD98 pKa = 3.54EE99 pKa = 4.27SAYY102 pKa = 9.67PGTGIGCEE110 pKa = 4.12GSYY113 pKa = 8.76TAPEE117 pKa = 4.24DD118 pKa = 3.55VAGNAFYY125 pKa = 11.11DD126 pKa = 4.06PVCDD130 pKa = 4.25VIAYY134 pKa = 9.42DD135 pKa = 3.9RR136 pKa = 11.84ALLQEE141 pKa = 4.59LSDD144 pKa = 3.93DD145 pKa = 3.72YY146 pKa = 11.89GRR148 pKa = 11.84FLVPVVMAHH157 pKa = 6.08EE158 pKa = 5.42FGHH161 pKa = 6.59AMQGRR166 pKa = 11.84FGFAEE171 pKa = 4.33SGRR174 pKa = 11.84SIQDD178 pKa = 2.99EE179 pKa = 4.39TQADD183 pKa = 4.26CLAGAWTRR191 pKa = 11.84WVADD195 pKa = 3.23GRR197 pKa = 11.84AAHH200 pKa = 6.64LSLRR204 pKa = 11.84EE205 pKa = 4.06PEE207 pKa = 4.32LDD209 pKa = 3.43DD210 pKa = 3.65VVRR213 pKa = 11.84GFLLLRR219 pKa = 11.84DD220 pKa = 4.48DD221 pKa = 4.65VGSDD225 pKa = 3.48PDD227 pKa = 3.9DD228 pKa = 4.26VEE230 pKa = 4.62AHH232 pKa = 5.93GSYY235 pKa = 10.53FDD237 pKa = 4.04RR238 pKa = 11.84VSAFYY243 pKa = 10.98DD244 pKa = 3.47GFEE247 pKa = 4.57RR248 pKa = 11.84GLGTCRR254 pKa = 11.84DD255 pKa = 3.59AFGEE259 pKa = 4.09DD260 pKa = 3.27RR261 pKa = 11.84LFTAAAFTPTDD272 pKa = 4.19LDD274 pKa = 3.3QGNADD279 pKa = 3.81FADD282 pKa = 3.4IVDD285 pKa = 3.93WVGATLPVFWGEE297 pKa = 3.94VFPAAFGGEE306 pKa = 4.17FEE308 pKa = 4.49EE309 pKa = 4.83PEE311 pKa = 3.65IAGFVGSAPGCDD323 pKa = 3.5GLDD326 pKa = 3.63GRR328 pKa = 11.84DD329 pKa = 3.64LGYY332 pKa = 10.72CADD335 pKa = 4.47DD336 pKa = 3.26RR337 pKa = 11.84TVYY340 pKa = 10.28IDD342 pKa = 3.58EE343 pKa = 4.59TDD345 pKa = 3.84LAVPAYY351 pKa = 10.53DD352 pKa = 4.39EE353 pKa = 4.8IGDD356 pKa = 3.96FALATAMALPYY367 pKa = 10.55ALAVRR372 pKa = 11.84DD373 pKa = 3.9QAGLPVDD380 pKa = 4.78DD381 pKa = 5.58GEE383 pKa = 4.44ATRR386 pKa = 11.84SAVCLTGWYY395 pKa = 8.42QAQWYY400 pKa = 8.51NAEE403 pKa = 3.99FSDD406 pKa = 4.64VVGAEE411 pKa = 4.07ISPGDD416 pKa = 4.05LDD418 pKa = 3.88EE419 pKa = 5.52AVQFLLRR426 pKa = 11.84YY427 pKa = 9.06GVEE430 pKa = 4.21DD431 pKa = 3.69EE432 pKa = 4.81VFPGTDD438 pKa = 2.71ASGFEE443 pKa = 4.14LVGAFRR449 pKa = 11.84NGFLDD454 pKa = 3.98GGAACGIGVV463 pKa = 3.7
Molecular weight: 49.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H7SHZ6|A0A1H7SHZ6_9ACTN Uncharacterized protein OS=Blastococcus sp. DSM 46786 OX=1798227 GN=SAMN04515665_11798 PE=4 SV=1
MM1 pKa = 7.44PRR3 pKa = 11.84VTVRR7 pKa = 11.84SVRR10 pKa = 11.84IARR13 pKa = 11.84SARR16 pKa = 11.84TARR19 pKa = 11.84RR20 pKa = 11.84GIVPAVTGRR29 pKa = 11.84TAVPRR34 pKa = 11.84VTVRR38 pKa = 11.84SVRR41 pKa = 11.84TARR44 pKa = 11.84RR45 pKa = 11.84VTGPAVTGRR54 pKa = 11.84TAVRR58 pKa = 3.48
MM1 pKa = 7.44PRR3 pKa = 11.84VTVRR7 pKa = 11.84SVRR10 pKa = 11.84IARR13 pKa = 11.84SARR16 pKa = 11.84TARR19 pKa = 11.84RR20 pKa = 11.84GIVPAVTGRR29 pKa = 11.84TAVPRR34 pKa = 11.84VTVRR38 pKa = 11.84SVRR41 pKa = 11.84TARR44 pKa = 11.84RR45 pKa = 11.84VTGPAVTGRR54 pKa = 11.84TAVRR58 pKa = 3.48
Molecular weight: 6.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1376897 |
28 |
2197 |
312.9 |
33.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.452 ± 0.058 | 0.685 ± 0.012 |
6.142 ± 0.03 | 5.728 ± 0.036 |
2.577 ± 0.025 | 9.825 ± 0.037 |
1.985 ± 0.018 | 2.804 ± 0.026 |
1.333 ± 0.022 | 10.758 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.611 ± 0.014 | 1.409 ± 0.017 |
6.28 ± 0.035 | 2.682 ± 0.021 |
8.205 ± 0.041 | 4.68 ± 0.023 |
5.863 ± 0.029 | 9.757 ± 0.038 |
1.485 ± 0.018 | 1.74 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
