
Olavius algarvensis Delta 1 endosymbiont
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; unclassified Deltaproteobacteria
Average proteome isoelectric point is 7.1
Get precalculated fractions of proteins
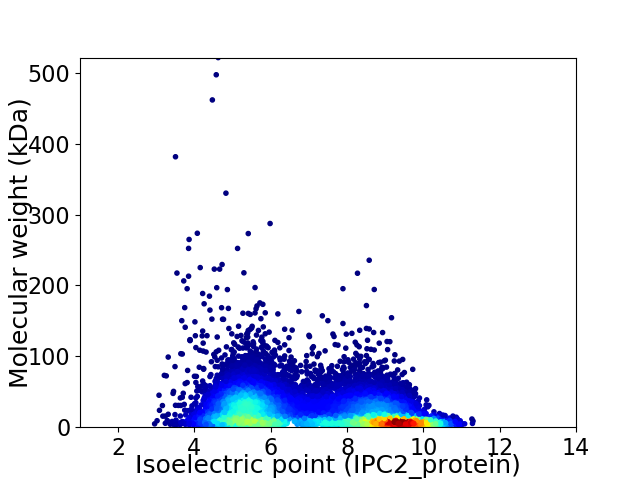
Virtual 2D-PAGE plot for 12909 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6J4XYF8|A0A6J4XYF8_9DELT Uncharacterized protein OS=Olavius algarvensis Delta 1 endosymbiont OX=334747 GN=D1AOALGA4SA_4813 PE=4 SV=1
MM1 pKa = 7.42GRR3 pKa = 11.84AKK5 pKa = 10.59QNLLQDD11 pKa = 3.99NPQQTEE17 pKa = 3.86VSRR20 pKa = 11.84TFLLFGDD27 pKa = 4.11PAMVLQVPLPRR38 pKa = 11.84RR39 pKa = 11.84PEE41 pKa = 3.95GLTAVLNADD50 pKa = 3.64TTVEE54 pKa = 4.32LNWDD58 pKa = 3.63SALDD62 pKa = 3.99ANGGAAAGYY71 pKa = 9.86NIYY74 pKa = 10.42RR75 pKa = 11.84GPWPNGPWTQLNDD88 pKa = 3.5TALMDD93 pKa = 3.94TVYY96 pKa = 10.98SDD98 pKa = 4.32SDD100 pKa = 3.83LTSGAAYY107 pKa = 9.91YY108 pKa = 10.28YY109 pKa = 11.07VVTALDD115 pKa = 3.69ADD117 pKa = 4.14GDD119 pKa = 3.99EE120 pKa = 4.97SVYY123 pKa = 10.97SDD125 pKa = 3.78PASAVLTDD133 pKa = 3.43SDD135 pKa = 4.76GDD137 pKa = 3.79GLYY140 pKa = 10.95DD141 pKa = 3.76LLEE144 pKa = 4.42AAGCTDD150 pKa = 3.63GNMSDD155 pKa = 3.74SDD157 pKa = 4.27GDD159 pKa = 3.98GLSDD163 pKa = 3.47GTEE166 pKa = 3.79DD167 pKa = 3.74ANRR170 pKa = 11.84NGTLDD175 pKa = 3.49AGEE178 pKa = 4.82TDD180 pKa = 3.64PCDD183 pKa = 3.86ADD185 pKa = 3.91SDD187 pKa = 4.83ADD189 pKa = 3.94TLPDD193 pKa = 3.07GWEE196 pKa = 3.86TANGFNALSGSGDD209 pKa = 3.71DD210 pKa = 5.75DD211 pKa = 4.37AAADD215 pKa = 4.06PDD217 pKa = 4.17GDD219 pKa = 3.1GWTNYY224 pKa = 8.77QEE226 pKa = 4.58YY227 pKa = 10.56LSCTDD232 pKa = 3.67PADD235 pKa = 3.95AASVPAAPTVPALNYY250 pKa = 7.33PANGSEE256 pKa = 4.17TTTLDD261 pKa = 2.85PWLSVSNAADD271 pKa = 4.15ADD273 pKa = 4.25CQDD276 pKa = 3.06LRR278 pKa = 11.84YY279 pKa = 10.34VFEE282 pKa = 5.15LYY284 pKa = 10.93ADD286 pKa = 4.1AGLTTLVDD294 pKa = 3.41ATAAEE299 pKa = 5.42GIAEE303 pKa = 4.46GDD305 pKa = 3.62NTTAWQPGSALNEE318 pKa = 3.6NAQYY322 pKa = 10.55YY323 pKa = 7.91WRR325 pKa = 11.84ARR327 pKa = 11.84AFDD330 pKa = 4.11SLASSDD336 pKa = 3.71WLATAAFFVNTTAEE350 pKa = 4.31APSTPAVSAPPDD362 pKa = 3.6GSEE365 pKa = 3.91VSVRR369 pKa = 11.84RR370 pKa = 11.84PSLEE374 pKa = 3.88VINAVDD380 pKa = 4.03ADD382 pKa = 4.28GDD384 pKa = 3.95PLTYY388 pKa = 10.75AFRR391 pKa = 11.84VYY393 pKa = 11.0ADD395 pKa = 4.05EE396 pKa = 4.23NLEE399 pKa = 4.19TLVDD403 pKa = 3.93SQSAVAEE410 pKa = 4.43GSGATTAWQVAAEE423 pKa = 4.45LDD425 pKa = 4.18DD426 pKa = 4.01NTDD429 pKa = 3.35YY430 pKa = 10.74WWTVQATDD438 pKa = 5.37DD439 pKa = 4.17EE440 pKa = 5.17DD441 pKa = 4.07LTGDD445 pKa = 3.15WSVADD450 pKa = 3.7RR451 pKa = 11.84FFVNTANDD459 pKa = 3.68APTTPPGGSPAIGAEE474 pKa = 4.15LATTTPLLAVVHH486 pKa = 5.93ATDD489 pKa = 4.39ADD491 pKa = 4.29LDD493 pKa = 3.85ALSYY497 pKa = 10.62FFEE500 pKa = 6.19IDD502 pKa = 2.87TAATFDD508 pKa = 4.05SFEE511 pKa = 4.74LEE513 pKa = 4.1QSDD516 pKa = 4.53EE517 pKa = 4.24LTEE520 pKa = 3.99EE521 pKa = 4.59AGVSTSWATQEE532 pKa = 5.65LIDD535 pKa = 3.59NTTYY539 pKa = 9.69YY540 pKa = 9.66WRR542 pKa = 11.84VRR544 pKa = 11.84AFDD547 pKa = 3.61GAAYY551 pKa = 9.64GPWHH555 pKa = 6.06NASFFVNTANDD566 pKa = 3.54APGAPTVDD574 pKa = 3.46HH575 pKa = 7.18PGDD578 pKa = 3.69GSEE581 pKa = 4.41VSTLQPTLTVKK592 pKa = 10.7AAIDD596 pKa = 3.66VDD598 pKa = 4.84LDD600 pKa = 3.52ALVYY604 pKa = 10.69DD605 pKa = 4.27FEE607 pKa = 6.12LYY609 pKa = 10.7EE610 pKa = 4.33DD611 pKa = 4.29TEE613 pKa = 4.41LTTLVTDD620 pKa = 3.74VAGVGLAWQVDD631 pKa = 3.76VTLDD635 pKa = 3.62DD636 pKa = 3.48NRR638 pKa = 11.84SYY640 pKa = 10.94FWRR643 pKa = 11.84ARR645 pKa = 11.84AVDD648 pKa = 3.37AHH650 pKa = 6.4GAAGPWSVLVSFFVNTANDD669 pKa = 4.0TPGTPVLNNPLSGGIATALTPTLSVFNADD698 pKa = 5.03DD699 pKa = 3.8TDD701 pKa = 3.92QDD703 pKa = 3.59ALTYY707 pKa = 10.81AFEE710 pKa = 4.95LYY712 pKa = 10.94ADD714 pKa = 4.49ADD716 pKa = 3.98LSQLVVGAFVAEE728 pKa = 4.19GHH730 pKa = 7.32LITSWTVSATLDD742 pKa = 4.14DD743 pKa = 3.74NGIYY747 pKa = 9.91YY748 pKa = 9.52WRR750 pKa = 11.84ARR752 pKa = 11.84ADD754 pKa = 3.93DD755 pKa = 4.37GQHH758 pKa = 6.39PGAWMPTAVLEE769 pKa = 4.02IHH771 pKa = 6.17TAGTNTAYY779 pKa = 10.12EE780 pKa = 4.35IVTQQPVSAAATARR794 pKa = 11.84QTVSVSAVDD803 pKa = 3.4HH804 pKa = 6.71PIYY807 pKa = 10.07NTVVEE812 pKa = 5.03LPPGALPRR820 pKa = 11.84DD821 pKa = 3.78CSINIGPVTNPPALPRR837 pKa = 11.84GARR840 pKa = 11.84KK841 pKa = 9.22IGHH844 pKa = 4.79VTEE847 pKa = 5.5FGPSGMTFNVPVVIKK862 pKa = 10.63LPYY865 pKa = 8.8TADD868 pKa = 3.43ALNQARR874 pKa = 11.84VANPVEE880 pKa = 4.33LSVYY884 pKa = 9.64WYY886 pKa = 10.24DD887 pKa = 3.48PSLLAWVPVEE897 pKa = 4.26ILDD900 pKa = 3.65IDD902 pKa = 4.8RR903 pKa = 11.84INKK906 pKa = 8.9LVSIEE911 pKa = 3.96TDD913 pKa = 2.98HH914 pKa = 7.65FSMYY918 pKa = 10.38TIGADD923 pKa = 3.26VADD926 pKa = 4.15ASGGGGGTCFIKK938 pKa = 10.45AVHH941 pKa = 6.12RR942 pKa = 11.84AEE944 pKa = 4.04TVFFIGNWSDD954 pKa = 3.17ISICGLLALVGLLWMGRR971 pKa = 11.84RR972 pKa = 11.84RR973 pKa = 11.84RR974 pKa = 11.84HH975 pKa = 4.81
MM1 pKa = 7.42GRR3 pKa = 11.84AKK5 pKa = 10.59QNLLQDD11 pKa = 3.99NPQQTEE17 pKa = 3.86VSRR20 pKa = 11.84TFLLFGDD27 pKa = 4.11PAMVLQVPLPRR38 pKa = 11.84RR39 pKa = 11.84PEE41 pKa = 3.95GLTAVLNADD50 pKa = 3.64TTVEE54 pKa = 4.32LNWDD58 pKa = 3.63SALDD62 pKa = 3.99ANGGAAAGYY71 pKa = 9.86NIYY74 pKa = 10.42RR75 pKa = 11.84GPWPNGPWTQLNDD88 pKa = 3.5TALMDD93 pKa = 3.94TVYY96 pKa = 10.98SDD98 pKa = 4.32SDD100 pKa = 3.83LTSGAAYY107 pKa = 9.91YY108 pKa = 10.28YY109 pKa = 11.07VVTALDD115 pKa = 3.69ADD117 pKa = 4.14GDD119 pKa = 3.99EE120 pKa = 4.97SVYY123 pKa = 10.97SDD125 pKa = 3.78PASAVLTDD133 pKa = 3.43SDD135 pKa = 4.76GDD137 pKa = 3.79GLYY140 pKa = 10.95DD141 pKa = 3.76LLEE144 pKa = 4.42AAGCTDD150 pKa = 3.63GNMSDD155 pKa = 3.74SDD157 pKa = 4.27GDD159 pKa = 3.98GLSDD163 pKa = 3.47GTEE166 pKa = 3.79DD167 pKa = 3.74ANRR170 pKa = 11.84NGTLDD175 pKa = 3.49AGEE178 pKa = 4.82TDD180 pKa = 3.64PCDD183 pKa = 3.86ADD185 pKa = 3.91SDD187 pKa = 4.83ADD189 pKa = 3.94TLPDD193 pKa = 3.07GWEE196 pKa = 3.86TANGFNALSGSGDD209 pKa = 3.71DD210 pKa = 5.75DD211 pKa = 4.37AAADD215 pKa = 4.06PDD217 pKa = 4.17GDD219 pKa = 3.1GWTNYY224 pKa = 8.77QEE226 pKa = 4.58YY227 pKa = 10.56LSCTDD232 pKa = 3.67PADD235 pKa = 3.95AASVPAAPTVPALNYY250 pKa = 7.33PANGSEE256 pKa = 4.17TTTLDD261 pKa = 2.85PWLSVSNAADD271 pKa = 4.15ADD273 pKa = 4.25CQDD276 pKa = 3.06LRR278 pKa = 11.84YY279 pKa = 10.34VFEE282 pKa = 5.15LYY284 pKa = 10.93ADD286 pKa = 4.1AGLTTLVDD294 pKa = 3.41ATAAEE299 pKa = 5.42GIAEE303 pKa = 4.46GDD305 pKa = 3.62NTTAWQPGSALNEE318 pKa = 3.6NAQYY322 pKa = 10.55YY323 pKa = 7.91WRR325 pKa = 11.84ARR327 pKa = 11.84AFDD330 pKa = 4.11SLASSDD336 pKa = 3.71WLATAAFFVNTTAEE350 pKa = 4.31APSTPAVSAPPDD362 pKa = 3.6GSEE365 pKa = 3.91VSVRR369 pKa = 11.84RR370 pKa = 11.84PSLEE374 pKa = 3.88VINAVDD380 pKa = 4.03ADD382 pKa = 4.28GDD384 pKa = 3.95PLTYY388 pKa = 10.75AFRR391 pKa = 11.84VYY393 pKa = 11.0ADD395 pKa = 4.05EE396 pKa = 4.23NLEE399 pKa = 4.19TLVDD403 pKa = 3.93SQSAVAEE410 pKa = 4.43GSGATTAWQVAAEE423 pKa = 4.45LDD425 pKa = 4.18DD426 pKa = 4.01NTDD429 pKa = 3.35YY430 pKa = 10.74WWTVQATDD438 pKa = 5.37DD439 pKa = 4.17EE440 pKa = 5.17DD441 pKa = 4.07LTGDD445 pKa = 3.15WSVADD450 pKa = 3.7RR451 pKa = 11.84FFVNTANDD459 pKa = 3.68APTTPPGGSPAIGAEE474 pKa = 4.15LATTTPLLAVVHH486 pKa = 5.93ATDD489 pKa = 4.39ADD491 pKa = 4.29LDD493 pKa = 3.85ALSYY497 pKa = 10.62FFEE500 pKa = 6.19IDD502 pKa = 2.87TAATFDD508 pKa = 4.05SFEE511 pKa = 4.74LEE513 pKa = 4.1QSDD516 pKa = 4.53EE517 pKa = 4.24LTEE520 pKa = 3.99EE521 pKa = 4.59AGVSTSWATQEE532 pKa = 5.65LIDD535 pKa = 3.59NTTYY539 pKa = 9.69YY540 pKa = 9.66WRR542 pKa = 11.84VRR544 pKa = 11.84AFDD547 pKa = 3.61GAAYY551 pKa = 9.64GPWHH555 pKa = 6.06NASFFVNTANDD566 pKa = 3.54APGAPTVDD574 pKa = 3.46HH575 pKa = 7.18PGDD578 pKa = 3.69GSEE581 pKa = 4.41VSTLQPTLTVKK592 pKa = 10.7AAIDD596 pKa = 3.66VDD598 pKa = 4.84LDD600 pKa = 3.52ALVYY604 pKa = 10.69DD605 pKa = 4.27FEE607 pKa = 6.12LYY609 pKa = 10.7EE610 pKa = 4.33DD611 pKa = 4.29TEE613 pKa = 4.41LTTLVTDD620 pKa = 3.74VAGVGLAWQVDD631 pKa = 3.76VTLDD635 pKa = 3.62DD636 pKa = 3.48NRR638 pKa = 11.84SYY640 pKa = 10.94FWRR643 pKa = 11.84ARR645 pKa = 11.84AVDD648 pKa = 3.37AHH650 pKa = 6.4GAAGPWSVLVSFFVNTANDD669 pKa = 4.0TPGTPVLNNPLSGGIATALTPTLSVFNADD698 pKa = 5.03DD699 pKa = 3.8TDD701 pKa = 3.92QDD703 pKa = 3.59ALTYY707 pKa = 10.81AFEE710 pKa = 4.95LYY712 pKa = 10.94ADD714 pKa = 4.49ADD716 pKa = 3.98LSQLVVGAFVAEE728 pKa = 4.19GHH730 pKa = 7.32LITSWTVSATLDD742 pKa = 4.14DD743 pKa = 3.74NGIYY747 pKa = 9.91YY748 pKa = 9.52WRR750 pKa = 11.84ARR752 pKa = 11.84ADD754 pKa = 3.93DD755 pKa = 4.37GQHH758 pKa = 6.39PGAWMPTAVLEE769 pKa = 4.02IHH771 pKa = 6.17TAGTNTAYY779 pKa = 10.12EE780 pKa = 4.35IVTQQPVSAAATARR794 pKa = 11.84QTVSVSAVDD803 pKa = 3.4HH804 pKa = 6.71PIYY807 pKa = 10.07NTVVEE812 pKa = 5.03LPPGALPRR820 pKa = 11.84DD821 pKa = 3.78CSINIGPVTNPPALPRR837 pKa = 11.84GARR840 pKa = 11.84KK841 pKa = 9.22IGHH844 pKa = 4.79VTEE847 pKa = 5.5FGPSGMTFNVPVVIKK862 pKa = 10.63LPYY865 pKa = 8.8TADD868 pKa = 3.43ALNQARR874 pKa = 11.84VANPVEE880 pKa = 4.33LSVYY884 pKa = 9.64WYY886 pKa = 10.24DD887 pKa = 3.48PSLLAWVPVEE897 pKa = 4.26ILDD900 pKa = 3.65IDD902 pKa = 4.8RR903 pKa = 11.84INKK906 pKa = 8.9LVSIEE911 pKa = 3.96TDD913 pKa = 2.98HH914 pKa = 7.65FSMYY918 pKa = 10.38TIGADD923 pKa = 3.26VADD926 pKa = 4.15ASGGGGGTCFIKK938 pKa = 10.45AVHH941 pKa = 6.12RR942 pKa = 11.84AEE944 pKa = 4.04TVFFIGNWSDD954 pKa = 3.17ISICGLLALVGLLWMGRR971 pKa = 11.84RR972 pKa = 11.84RR973 pKa = 11.84RR974 pKa = 11.84HH975 pKa = 4.81
Molecular weight: 103.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6J4YHL3|A0A6J4YHL3_9DELT Oxidoreductase short-chain dehydrogenase/reductase family OS=Olavius algarvensis Delta 1 endosymbiont OX=334747 GN=D1AOALGA4SA_9704 PE=4 SV=1
QQ1 pKa = 7.1PATRR5 pKa = 11.84NPQRR9 pKa = 11.84ATRR12 pKa = 11.84NPQRR16 pKa = 11.84ATRR19 pKa = 11.84NPQPATRR26 pKa = 11.84NAQPATRR33 pKa = 11.84NAQRR37 pKa = 11.84ATRR40 pKa = 11.84NPNPQLATRR49 pKa = 11.84NPQPVTRR56 pKa = 11.84NPKK59 pKa = 8.86PATRR63 pKa = 11.84NPQSAIRR70 pKa = 11.84NPHH73 pKa = 5.43SAIHH77 pKa = 5.67SS78 pKa = 3.82
QQ1 pKa = 7.1PATRR5 pKa = 11.84NPQRR9 pKa = 11.84ATRR12 pKa = 11.84NPQRR16 pKa = 11.84ATRR19 pKa = 11.84NPQPATRR26 pKa = 11.84NAQPATRR33 pKa = 11.84NAQRR37 pKa = 11.84ATRR40 pKa = 11.84NPNPQLATRR49 pKa = 11.84NPQPVTRR56 pKa = 11.84NPKK59 pKa = 8.86PATRR63 pKa = 11.84NPQSAIRR70 pKa = 11.84NPHH73 pKa = 5.43SAIHH77 pKa = 5.67SS78 pKa = 3.82
Molecular weight: 8.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2800634 |
20 |
4859 |
217.0 |
24.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.561 ± 0.029 | 1.255 ± 0.012 |
5.574 ± 0.026 | 5.955 ± 0.022 |
4.46 ± 0.018 | 7.331 ± 0.023 |
2.069 ± 0.011 | 6.964 ± 0.023 |
5.434 ± 0.029 | 9.868 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.619 ± 0.014 | 3.846 ± 0.02 |
4.515 ± 0.018 | 3.694 ± 0.016 |
5.869 ± 0.024 | 6.029 ± 0.02 |
4.96 ± 0.024 | 6.659 ± 0.019 |
1.238 ± 0.01 | 3.101 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
