
Bacillus sp. FJAT-27916
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; unclassified Bacillus (in: Bacteria)
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
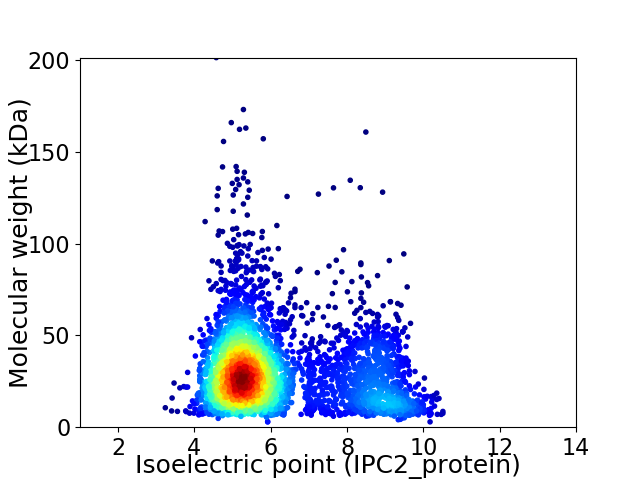
Virtual 2D-PAGE plot for 3770 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K9GIL8|A0A0K9GIL8_9BACI Elongation factor Tu OS=Bacillus sp. FJAT-27916 OX=1679169 GN=tuf PE=3 SV=1
MM1 pKa = 7.52QIYY4 pKa = 9.96CSKK7 pKa = 10.43CGEE10 pKa = 4.16PFSRR14 pKa = 11.84QPRR17 pKa = 11.84YY18 pKa = 10.18CSACGNKK25 pKa = 7.92TEE27 pKa = 4.56RR28 pKa = 11.84KK29 pKa = 8.02SKK31 pKa = 10.64KK32 pKa = 9.45IIIWGLALITTLCLCVMASYY52 pKa = 11.07GFYY55 pKa = 10.76LFNEE59 pKa = 4.39KK60 pKa = 10.36KK61 pKa = 10.05EE62 pKa = 4.22AEE64 pKa = 4.01QSIQDD69 pKa = 3.31KK70 pKa = 10.23PEE72 pKa = 3.76QVKK75 pKa = 10.12EE76 pKa = 4.23VVKK79 pKa = 9.53KK80 pKa = 8.93TKK82 pKa = 10.52KK83 pKa = 9.28EE84 pKa = 3.97APKK87 pKa = 10.0PALQKK92 pKa = 10.02TSTAPKK98 pKa = 9.86DD99 pKa = 3.41VSQIIEE105 pKa = 3.96EE106 pKa = 4.28SLSKK110 pKa = 11.03VFTISNGTSQGSGFLINKK128 pKa = 9.71NGDD131 pKa = 3.31ILTNAHH137 pKa = 4.85VVEE140 pKa = 4.83GNTLAIVKK148 pKa = 9.25DD149 pKa = 3.93HH150 pKa = 6.94NGTEE154 pKa = 4.19HH155 pKa = 6.24QGTVIGYY162 pKa = 9.32SNEE165 pKa = 3.33VDD167 pKa = 2.81IALIRR172 pKa = 11.84VPNLANQTPLSIEE185 pKa = 4.27TTDD188 pKa = 3.29KK189 pKa = 11.27SQLGDD194 pKa = 3.74EE195 pKa = 4.94IIALGSPRR203 pKa = 11.84GLEE206 pKa = 3.92NSATLGNISGVDD218 pKa = 3.12RR219 pKa = 11.84TFVIEE224 pKa = 3.6PHH226 pKa = 6.72LYY228 pKa = 10.55DD229 pKa = 5.14GIYY232 pKa = 10.08QISAPLAPGSSGGPLLDD249 pKa = 3.58RR250 pKa = 11.84KK251 pKa = 7.48TEE253 pKa = 3.91KK254 pKa = 10.9VIAINSAKK262 pKa = 10.2MNGEE266 pKa = 4.06DD267 pKa = 4.33NIGFSIPLYY276 pKa = 10.65KK277 pKa = 10.23IMNTVTQWSSAPMSEE292 pKa = 4.6QEE294 pKa = 4.35INQLFYY300 pKa = 11.53NEE302 pKa = 4.83AGVFYY307 pKa = 11.37YY308 pKa = 10.66EE309 pKa = 4.65DD310 pKa = 4.09LYY312 pKa = 11.74GNEE315 pKa = 4.96GYY317 pKa = 10.61FDD319 pKa = 3.79GGSYY323 pKa = 10.19EE324 pKa = 4.24EE325 pKa = 5.43EE326 pKa = 3.85YY327 pKa = 11.09DD328 pKa = 4.41SYY330 pKa = 11.85YY331 pKa = 10.96EE332 pKa = 4.39DD333 pKa = 3.99YY334 pKa = 10.8EE335 pKa = 4.17YY336 pKa = 10.62DD337 pKa = 3.64YY338 pKa = 11.59DD339 pKa = 4.68DD340 pKa = 5.2SEE342 pKa = 4.73FEE344 pKa = 4.07EE345 pKa = 5.84DD346 pKa = 3.39YY347 pKa = 11.44SEE349 pKa = 4.61YY350 pKa = 10.95DD351 pKa = 3.52DD352 pKa = 5.31YY353 pKa = 11.71EE354 pKa = 4.6YY355 pKa = 11.53DD356 pKa = 3.4HH357 pKa = 7.57DD358 pKa = 4.22EE359 pKa = 5.48GYY361 pKa = 11.29DD362 pKa = 3.88DD363 pKa = 5.38MYY365 pKa = 11.58DD366 pKa = 3.6DD367 pKa = 3.65SDD369 pKa = 4.46YY370 pKa = 11.67YY371 pKa = 11.09EE372 pKa = 4.91EE373 pKa = 5.85DD374 pKa = 3.68PEE376 pKa = 4.82EE377 pKa = 5.24DD378 pKa = 3.52DD379 pKa = 4.41SLMIEE384 pKa = 4.3EE385 pKa = 5.14DD386 pKa = 3.57SEE388 pKa = 4.11EE389 pKa = 5.14AYY391 pKa = 10.81DD392 pKa = 3.73DD393 pKa = 4.03TLVEE397 pKa = 4.07EE398 pKa = 4.76SDD400 pKa = 4.44LEE402 pKa = 4.32EE403 pKa = 5.02EE404 pKa = 4.95NDD406 pKa = 4.28DD407 pKa = 5.67DD408 pKa = 6.17DD409 pKa = 6.59IEE411 pKa = 4.21MDD413 pKa = 3.58PYY415 pKa = 11.33GNKK418 pKa = 9.88EE419 pKa = 4.12DD420 pKa = 3.9VDD422 pKa = 4.63SDD424 pKa = 4.15VSDD427 pKa = 5.49DD428 pKa = 4.57YY429 pKa = 11.95LDD431 pKa = 3.36QQ432 pKa = 5.83
MM1 pKa = 7.52QIYY4 pKa = 9.96CSKK7 pKa = 10.43CGEE10 pKa = 4.16PFSRR14 pKa = 11.84QPRR17 pKa = 11.84YY18 pKa = 10.18CSACGNKK25 pKa = 7.92TEE27 pKa = 4.56RR28 pKa = 11.84KK29 pKa = 8.02SKK31 pKa = 10.64KK32 pKa = 9.45IIIWGLALITTLCLCVMASYY52 pKa = 11.07GFYY55 pKa = 10.76LFNEE59 pKa = 4.39KK60 pKa = 10.36KK61 pKa = 10.05EE62 pKa = 4.22AEE64 pKa = 4.01QSIQDD69 pKa = 3.31KK70 pKa = 10.23PEE72 pKa = 3.76QVKK75 pKa = 10.12EE76 pKa = 4.23VVKK79 pKa = 9.53KK80 pKa = 8.93TKK82 pKa = 10.52KK83 pKa = 9.28EE84 pKa = 3.97APKK87 pKa = 10.0PALQKK92 pKa = 10.02TSTAPKK98 pKa = 9.86DD99 pKa = 3.41VSQIIEE105 pKa = 3.96EE106 pKa = 4.28SLSKK110 pKa = 11.03VFTISNGTSQGSGFLINKK128 pKa = 9.71NGDD131 pKa = 3.31ILTNAHH137 pKa = 4.85VVEE140 pKa = 4.83GNTLAIVKK148 pKa = 9.25DD149 pKa = 3.93HH150 pKa = 6.94NGTEE154 pKa = 4.19HH155 pKa = 6.24QGTVIGYY162 pKa = 9.32SNEE165 pKa = 3.33VDD167 pKa = 2.81IALIRR172 pKa = 11.84VPNLANQTPLSIEE185 pKa = 4.27TTDD188 pKa = 3.29KK189 pKa = 11.27SQLGDD194 pKa = 3.74EE195 pKa = 4.94IIALGSPRR203 pKa = 11.84GLEE206 pKa = 3.92NSATLGNISGVDD218 pKa = 3.12RR219 pKa = 11.84TFVIEE224 pKa = 3.6PHH226 pKa = 6.72LYY228 pKa = 10.55DD229 pKa = 5.14GIYY232 pKa = 10.08QISAPLAPGSSGGPLLDD249 pKa = 3.58RR250 pKa = 11.84KK251 pKa = 7.48TEE253 pKa = 3.91KK254 pKa = 10.9VIAINSAKK262 pKa = 10.2MNGEE266 pKa = 4.06DD267 pKa = 4.33NIGFSIPLYY276 pKa = 10.65KK277 pKa = 10.23IMNTVTQWSSAPMSEE292 pKa = 4.6QEE294 pKa = 4.35INQLFYY300 pKa = 11.53NEE302 pKa = 4.83AGVFYY307 pKa = 11.37YY308 pKa = 10.66EE309 pKa = 4.65DD310 pKa = 4.09LYY312 pKa = 11.74GNEE315 pKa = 4.96GYY317 pKa = 10.61FDD319 pKa = 3.79GGSYY323 pKa = 10.19EE324 pKa = 4.24EE325 pKa = 5.43EE326 pKa = 3.85YY327 pKa = 11.09DD328 pKa = 4.41SYY330 pKa = 11.85YY331 pKa = 10.96EE332 pKa = 4.39DD333 pKa = 3.99YY334 pKa = 10.8EE335 pKa = 4.17YY336 pKa = 10.62DD337 pKa = 3.64YY338 pKa = 11.59DD339 pKa = 4.68DD340 pKa = 5.2SEE342 pKa = 4.73FEE344 pKa = 4.07EE345 pKa = 5.84DD346 pKa = 3.39YY347 pKa = 11.44SEE349 pKa = 4.61YY350 pKa = 10.95DD351 pKa = 3.52DD352 pKa = 5.31YY353 pKa = 11.71EE354 pKa = 4.6YY355 pKa = 11.53DD356 pKa = 3.4HH357 pKa = 7.57DD358 pKa = 4.22EE359 pKa = 5.48GYY361 pKa = 11.29DD362 pKa = 3.88DD363 pKa = 5.38MYY365 pKa = 11.58DD366 pKa = 3.6DD367 pKa = 3.65SDD369 pKa = 4.46YY370 pKa = 11.67YY371 pKa = 11.09EE372 pKa = 4.91EE373 pKa = 5.85DD374 pKa = 3.68PEE376 pKa = 4.82EE377 pKa = 5.24DD378 pKa = 3.52DD379 pKa = 4.41SLMIEE384 pKa = 4.3EE385 pKa = 5.14DD386 pKa = 3.57SEE388 pKa = 4.11EE389 pKa = 5.14AYY391 pKa = 10.81DD392 pKa = 3.73DD393 pKa = 4.03TLVEE397 pKa = 4.07EE398 pKa = 4.76SDD400 pKa = 4.44LEE402 pKa = 4.32EE403 pKa = 5.02EE404 pKa = 4.95NDD406 pKa = 4.28DD407 pKa = 5.67DD408 pKa = 6.17DD409 pKa = 6.59IEE411 pKa = 4.21MDD413 pKa = 3.58PYY415 pKa = 11.33GNKK418 pKa = 9.88EE419 pKa = 4.12DD420 pKa = 3.9VDD422 pKa = 4.63SDD424 pKa = 4.15VSDD427 pKa = 5.49DD428 pKa = 4.57YY429 pKa = 11.95LDD431 pKa = 3.36QQ432 pKa = 5.83
Molecular weight: 48.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K9GBR2|A0A0K9GBR2_9BACI Nucleoside diphosphate kinase OS=Bacillus sp. FJAT-27916 OX=1679169 GN=ndk PE=3 SV=1
MM1 pKa = 7.26QGGEE5 pKa = 4.35EE6 pKa = 4.34PCGFPAIDD14 pKa = 3.83YY15 pKa = 8.33VWSGRR20 pKa = 11.84RR21 pKa = 11.84KK22 pKa = 9.03GKK24 pKa = 10.68GMVEE28 pKa = 3.77RR29 pKa = 11.84WEE31 pKa = 4.48MPCSSRR37 pKa = 11.84LFPGQGIYY45 pKa = 10.19RR46 pKa = 11.84AVEE49 pKa = 3.62GWEE52 pKa = 3.77RR53 pKa = 11.84GGRR56 pKa = 11.84SHH58 pKa = 7.27AVPTQCPYY66 pKa = 10.65KK67 pKa = 10.81GYY69 pKa = 10.24IGWGKK74 pKa = 10.2GGHH77 pKa = 5.31RR78 pKa = 11.84QRR80 pKa = 11.84KK81 pKa = 5.67PTQCPGKK88 pKa = 10.74GYY90 pKa = 10.23IGWGKK95 pKa = 10.26GGHH98 pKa = 5.24RR99 pKa = 11.84QRR101 pKa = 11.84NPIQCPGKK109 pKa = 10.82GYY111 pKa = 10.29IGWGKK116 pKa = 10.2GGHH119 pKa = 5.31RR120 pKa = 11.84QRR122 pKa = 11.84KK123 pKa = 5.67PTQCPGKK130 pKa = 10.74GYY132 pKa = 10.14IGWGKK137 pKa = 10.58GEE139 pKa = 3.85HH140 pKa = 5.8RR141 pKa = 11.84QRR143 pKa = 11.84KK144 pKa = 6.12PTQCPGKK151 pKa = 10.74GYY153 pKa = 10.23IGWGKK158 pKa = 10.2GGHH161 pKa = 5.31RR162 pKa = 11.84QRR164 pKa = 11.84KK165 pKa = 5.95PTQCPYY171 pKa = 10.48KK172 pKa = 10.77GYY174 pKa = 10.24IGWGKK179 pKa = 10.26GGHH182 pKa = 5.24RR183 pKa = 11.84QRR185 pKa = 11.84NPIQCPGKK193 pKa = 10.73GYY195 pKa = 10.27IGG197 pKa = 3.56
MM1 pKa = 7.26QGGEE5 pKa = 4.35EE6 pKa = 4.34PCGFPAIDD14 pKa = 3.83YY15 pKa = 8.33VWSGRR20 pKa = 11.84RR21 pKa = 11.84KK22 pKa = 9.03GKK24 pKa = 10.68GMVEE28 pKa = 3.77RR29 pKa = 11.84WEE31 pKa = 4.48MPCSSRR37 pKa = 11.84LFPGQGIYY45 pKa = 10.19RR46 pKa = 11.84AVEE49 pKa = 3.62GWEE52 pKa = 3.77RR53 pKa = 11.84GGRR56 pKa = 11.84SHH58 pKa = 7.27AVPTQCPYY66 pKa = 10.65KK67 pKa = 10.81GYY69 pKa = 10.24IGWGKK74 pKa = 10.2GGHH77 pKa = 5.31RR78 pKa = 11.84QRR80 pKa = 11.84KK81 pKa = 5.67PTQCPGKK88 pKa = 10.74GYY90 pKa = 10.23IGWGKK95 pKa = 10.26GGHH98 pKa = 5.24RR99 pKa = 11.84QRR101 pKa = 11.84NPIQCPGKK109 pKa = 10.82GYY111 pKa = 10.29IGWGKK116 pKa = 10.2GGHH119 pKa = 5.31RR120 pKa = 11.84QRR122 pKa = 11.84KK123 pKa = 5.67PTQCPGKK130 pKa = 10.74GYY132 pKa = 10.14IGWGKK137 pKa = 10.58GEE139 pKa = 3.85HH140 pKa = 5.8RR141 pKa = 11.84QRR143 pKa = 11.84KK144 pKa = 6.12PTQCPGKK151 pKa = 10.74GYY153 pKa = 10.23IGWGKK158 pKa = 10.2GGHH161 pKa = 5.31RR162 pKa = 11.84QRR164 pKa = 11.84KK165 pKa = 5.95PTQCPYY171 pKa = 10.48KK172 pKa = 10.77GYY174 pKa = 10.24IGWGKK179 pKa = 10.26GGHH182 pKa = 5.24RR183 pKa = 11.84QRR185 pKa = 11.84NPIQCPGKK193 pKa = 10.73GYY195 pKa = 10.27IGG197 pKa = 3.56
Molecular weight: 21.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1089385 |
26 |
1873 |
289.0 |
32.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.484 ± 0.041 | 0.75 ± 0.012 |
5.144 ± 0.037 | 7.737 ± 0.048 |
4.377 ± 0.039 | 7.207 ± 0.039 |
2.072 ± 0.021 | 7.79 ± 0.038 |
6.738 ± 0.04 | 9.716 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.991 ± 0.019 | 4.023 ± 0.022 |
3.604 ± 0.022 | 3.495 ± 0.026 |
4.254 ± 0.035 | 5.917 ± 0.032 |
5.282 ± 0.03 | 6.733 ± 0.032 |
1.033 ± 0.016 | 3.653 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
