
Mycobacterium grossiae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Mycobacteriaceae; Mycobacterium
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
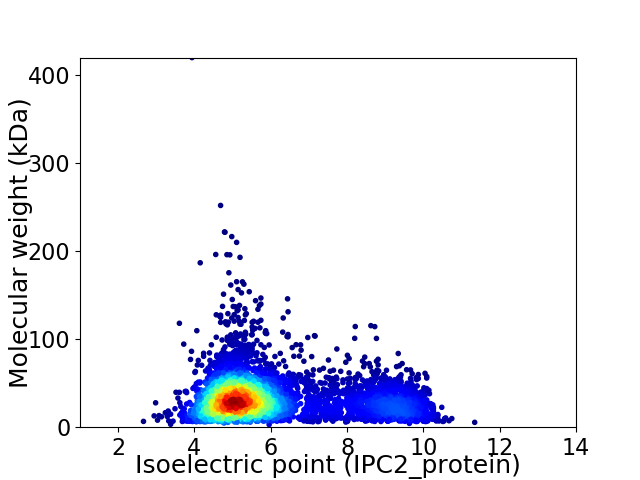
Virtual 2D-PAGE plot for 5542 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E8Q0S5|A0A1E8Q0S5_9MYCO Protein rarD OS=Mycobacterium grossiae OX=1552759 GN=BEL07_22625 PE=3 SV=1
MM1 pKa = 7.2NVDD4 pKa = 3.36SSGGAITSEE13 pKa = 4.16HH14 pKa = 6.08RR15 pKa = 11.84TSDD18 pKa = 3.51DD19 pKa = 2.98TSAPTTVSQPAPTTDD34 pKa = 3.39SQSVSPAVSPAVSQPQDD51 pKa = 3.26APAAPASAPAKK62 pKa = 10.49NDD64 pKa = 3.37DD65 pKa = 4.18PAPAPTARR73 pKa = 11.84PAAPGAVVHH82 pKa = 7.01ADD84 pKa = 3.6TVSVGSTDD92 pKa = 3.36ATAVAPPPAAAVVSVPAASLDD113 pKa = 3.55ATAGSVASEE122 pKa = 4.39PEE124 pKa = 4.12SAALQQEE131 pKa = 4.59STVVSVLAAAVAPFVLPGPATPAASPLLWAVLAWTRR167 pKa = 11.84RR168 pKa = 11.84EE169 pKa = 4.1AEE171 pKa = 3.79VDD173 pKa = 3.57GSSEE177 pKa = 4.23TLRR180 pKa = 11.84TTAAAAPADD189 pKa = 3.92FEE191 pKa = 4.66VEE193 pKa = 3.52EE194 pKa = 4.4DD195 pKa = 3.51QPLFGNVLDD204 pKa = 4.43LHH206 pKa = 6.86PDD208 pKa = 3.44VTPGTVTVGDD218 pKa = 4.06GTAPAHH224 pKa = 5.84GTLTLDD230 pKa = 3.31GSGNFVYY237 pKa = 10.38TPDD240 pKa = 3.6QDD242 pKa = 4.69FSGDD246 pKa = 3.62DD247 pKa = 3.23QFGYY251 pKa = 9.24TLTDD255 pKa = 3.53FDD257 pKa = 4.62GVVVTATARR266 pKa = 11.84ITVTPAPDD274 pKa = 3.3VPVGSDD280 pKa = 2.72ADD282 pKa = 3.63VRR284 pKa = 11.84TDD286 pKa = 3.19EE287 pKa = 4.71DD288 pKa = 4.27TPITSTLPAAGDD300 pKa = 3.45VDD302 pKa = 4.25GDD304 pKa = 3.87EE305 pKa = 4.75LVYY308 pKa = 10.81AVAQSPQSGTVVVAADD324 pKa = 3.54GAYY327 pKa = 9.4TYY329 pKa = 10.55TPAPNFYY336 pKa = 10.62GVDD339 pKa = 3.27SFGYY343 pKa = 7.91TVSDD347 pKa = 3.46GTFTVGYY354 pKa = 9.26VVTVTVAPVDD364 pKa = 4.07DD365 pKa = 5.16APVALDD371 pKa = 4.24DD372 pKa = 4.39SVSGPEE378 pKa = 3.84DD379 pKa = 3.5SEE381 pKa = 4.42FTGVVPVTEE390 pKa = 3.92VDD392 pKa = 3.65GDD394 pKa = 3.93VLTYY398 pKa = 10.72GIDD401 pKa = 3.09RR402 pKa = 11.84GPQHH406 pKa = 6.07GTVTLGADD414 pKa = 3.18GRR416 pKa = 11.84YY417 pKa = 9.82VYY419 pKa = 10.47TPAADD424 pKa = 3.47FHH426 pKa = 6.58GADD429 pKa = 3.25SFTYY433 pKa = 9.52TVTDD437 pKa = 3.62GTTAPSTGTIAFTVTPVNDD456 pKa = 4.07PPRR459 pKa = 11.84SEE461 pKa = 4.53HH462 pKa = 6.8LDD464 pKa = 3.26VSTAEE469 pKa = 3.99DD470 pKa = 3.57TPYY473 pKa = 11.3DD474 pKa = 3.76GVLPTSDD481 pKa = 4.37PDD483 pKa = 4.27GDD485 pKa = 4.14PLTYY489 pKa = 10.73DD490 pKa = 3.82LFGTPVHH497 pKa = 5.74GTVIIRR503 pKa = 11.84PDD505 pKa = 3.06GSFTYY510 pKa = 10.47RR511 pKa = 11.84PDD513 pKa = 3.33ANYY516 pKa = 11.0NGADD520 pKa = 3.3EE521 pKa = 4.45FTYY524 pKa = 10.76HH525 pKa = 7.41VMDD528 pKa = 3.99GTTYY532 pKa = 8.39GQYY535 pKa = 10.22VVRR538 pKa = 11.84ITVTAVNDD546 pKa = 3.78VPVTTPAAFTTGPGVAVTDD565 pKa = 3.95TVAWHH570 pKa = 6.63ASDD573 pKa = 4.78VDD575 pKa = 3.98VADD578 pKa = 4.25EE579 pKa = 4.33LTFTAAAPTAGTVTMGADD597 pKa = 3.52GQFTYY602 pKa = 10.19TPRR605 pKa = 11.84PGFSGTDD612 pKa = 3.04TFVFTVTDD620 pKa = 3.64TAGATATGVVSVTVANQQPVAYY642 pKa = 9.58DD643 pKa = 3.19DD644 pKa = 5.28GYY646 pKa = 11.47QVTPGATLTVDD657 pKa = 3.41AAHH660 pKa = 6.97GLLANDD666 pKa = 4.32VDD668 pKa = 4.53PGDD671 pKa = 4.37PLTVTSVTQPTHH683 pKa = 4.79GTVTVAGDD691 pKa = 3.47GSFTYY696 pKa = 10.21TPTAGFSGDD705 pKa = 3.2DD706 pKa = 3.29WFSYY710 pKa = 10.33TITDD714 pKa = 3.68SAGATATATVFFGVAPASNLPPIAPDD740 pKa = 2.96WTLTTEE746 pKa = 4.17ADD748 pKa = 3.14VDD750 pKa = 4.13VTFDD754 pKa = 3.78PLAGVTDD761 pKa = 4.33PNGDD765 pKa = 3.92VLWVRR770 pKa = 11.84DD771 pKa = 4.11GEE773 pKa = 4.43QPAHH777 pKa = 6.65GFVQQYY783 pKa = 9.57TNGTIAYY790 pKa = 8.86VPDD793 pKa = 3.62GTFVGTDD800 pKa = 2.89GFTYY804 pKa = 10.05EE805 pKa = 4.21VHH807 pKa = 7.5DD808 pKa = 3.96GTNVVTVRR816 pKa = 11.84VTVITSPSTAPVATGDD832 pKa = 3.83EE833 pKa = 4.31VSMPEE838 pKa = 4.21GGTVTIDD845 pKa = 3.24PLANDD850 pKa = 4.21TFIGTPTVEE859 pKa = 5.05LISTAQYY866 pKa = 8.99GTVTVDD872 pKa = 2.77AATGKK877 pKa = 8.53LTYY880 pKa = 10.44VATAGTNALGDD891 pKa = 3.71SFAYY895 pKa = 10.29QITDD899 pKa = 2.9EE900 pKa = 4.2RR901 pKa = 11.84GRR903 pKa = 11.84WSVASVDD910 pKa = 3.34ITFTAGNRR918 pKa = 11.84PPVATDD924 pKa = 3.85DD925 pKa = 3.96VAATPIGTPVSIDD938 pKa = 3.44VLANDD943 pKa = 4.08YY944 pKa = 11.44DD945 pKa = 4.85RR946 pKa = 11.84DD947 pKa = 3.97GTTLTVSVVSQPEE960 pKa = 3.88FGGTAIARR968 pKa = 11.84KK969 pKa = 10.07DD970 pKa = 3.36GTIDD974 pKa = 3.49FTPTKK979 pKa = 9.41GYY981 pKa = 11.04VGMASFSYY989 pKa = 10.42EE990 pKa = 3.95VTDD993 pKa = 3.51EE994 pKa = 4.24SGAVARR1000 pKa = 11.84ATVTVTVYY1008 pKa = 11.11DD1009 pKa = 4.18PTPPDD1014 pKa = 3.93APVKK1018 pKa = 10.51SYY1020 pKa = 9.76TVGKK1024 pKa = 10.42GRR1026 pKa = 11.84TLTVSLGDD1034 pKa = 3.65GVLAGSGYY1042 pKa = 10.18QGWLTGLEE1050 pKa = 4.08TGPAHH1055 pKa = 6.33GVLRR1059 pKa = 11.84LAEE1062 pKa = 5.09DD1063 pKa = 4.02GSFTYY1068 pKa = 9.86TPRR1071 pKa = 11.84EE1072 pKa = 4.09GFVGTDD1078 pKa = 2.89SFVFYY1083 pKa = 10.99VHH1085 pKa = 6.24TRR1087 pKa = 11.84EE1088 pKa = 4.23QGDD1091 pKa = 4.26GPFTATITVTEE1102 pKa = 5.25DD1103 pKa = 4.04GPGTPPNDD1111 pKa = 3.4AVAVALSSSDD1121 pKa = 3.28TAYY1124 pKa = 8.34GTCGGDD1130 pKa = 2.8WFAGRR1135 pKa = 11.84DD1136 pKa = 3.95DD1137 pKa = 5.0PASTWPTCLFLGRR1150 pKa = 11.84PEE1152 pKa = 4.31DD1153 pKa = 3.65VV1154 pKa = 3.47
MM1 pKa = 7.2NVDD4 pKa = 3.36SSGGAITSEE13 pKa = 4.16HH14 pKa = 6.08RR15 pKa = 11.84TSDD18 pKa = 3.51DD19 pKa = 2.98TSAPTTVSQPAPTTDD34 pKa = 3.39SQSVSPAVSPAVSQPQDD51 pKa = 3.26APAAPASAPAKK62 pKa = 10.49NDD64 pKa = 3.37DD65 pKa = 4.18PAPAPTARR73 pKa = 11.84PAAPGAVVHH82 pKa = 7.01ADD84 pKa = 3.6TVSVGSTDD92 pKa = 3.36ATAVAPPPAAAVVSVPAASLDD113 pKa = 3.55ATAGSVASEE122 pKa = 4.39PEE124 pKa = 4.12SAALQQEE131 pKa = 4.59STVVSVLAAAVAPFVLPGPATPAASPLLWAVLAWTRR167 pKa = 11.84RR168 pKa = 11.84EE169 pKa = 4.1AEE171 pKa = 3.79VDD173 pKa = 3.57GSSEE177 pKa = 4.23TLRR180 pKa = 11.84TTAAAAPADD189 pKa = 3.92FEE191 pKa = 4.66VEE193 pKa = 3.52EE194 pKa = 4.4DD195 pKa = 3.51QPLFGNVLDD204 pKa = 4.43LHH206 pKa = 6.86PDD208 pKa = 3.44VTPGTVTVGDD218 pKa = 4.06GTAPAHH224 pKa = 5.84GTLTLDD230 pKa = 3.31GSGNFVYY237 pKa = 10.38TPDD240 pKa = 3.6QDD242 pKa = 4.69FSGDD246 pKa = 3.62DD247 pKa = 3.23QFGYY251 pKa = 9.24TLTDD255 pKa = 3.53FDD257 pKa = 4.62GVVVTATARR266 pKa = 11.84ITVTPAPDD274 pKa = 3.3VPVGSDD280 pKa = 2.72ADD282 pKa = 3.63VRR284 pKa = 11.84TDD286 pKa = 3.19EE287 pKa = 4.71DD288 pKa = 4.27TPITSTLPAAGDD300 pKa = 3.45VDD302 pKa = 4.25GDD304 pKa = 3.87EE305 pKa = 4.75LVYY308 pKa = 10.81AVAQSPQSGTVVVAADD324 pKa = 3.54GAYY327 pKa = 9.4TYY329 pKa = 10.55TPAPNFYY336 pKa = 10.62GVDD339 pKa = 3.27SFGYY343 pKa = 7.91TVSDD347 pKa = 3.46GTFTVGYY354 pKa = 9.26VVTVTVAPVDD364 pKa = 4.07DD365 pKa = 5.16APVALDD371 pKa = 4.24DD372 pKa = 4.39SVSGPEE378 pKa = 3.84DD379 pKa = 3.5SEE381 pKa = 4.42FTGVVPVTEE390 pKa = 3.92VDD392 pKa = 3.65GDD394 pKa = 3.93VLTYY398 pKa = 10.72GIDD401 pKa = 3.09RR402 pKa = 11.84GPQHH406 pKa = 6.07GTVTLGADD414 pKa = 3.18GRR416 pKa = 11.84YY417 pKa = 9.82VYY419 pKa = 10.47TPAADD424 pKa = 3.47FHH426 pKa = 6.58GADD429 pKa = 3.25SFTYY433 pKa = 9.52TVTDD437 pKa = 3.62GTTAPSTGTIAFTVTPVNDD456 pKa = 4.07PPRR459 pKa = 11.84SEE461 pKa = 4.53HH462 pKa = 6.8LDD464 pKa = 3.26VSTAEE469 pKa = 3.99DD470 pKa = 3.57TPYY473 pKa = 11.3DD474 pKa = 3.76GVLPTSDD481 pKa = 4.37PDD483 pKa = 4.27GDD485 pKa = 4.14PLTYY489 pKa = 10.73DD490 pKa = 3.82LFGTPVHH497 pKa = 5.74GTVIIRR503 pKa = 11.84PDD505 pKa = 3.06GSFTYY510 pKa = 10.47RR511 pKa = 11.84PDD513 pKa = 3.33ANYY516 pKa = 11.0NGADD520 pKa = 3.3EE521 pKa = 4.45FTYY524 pKa = 10.76HH525 pKa = 7.41VMDD528 pKa = 3.99GTTYY532 pKa = 8.39GQYY535 pKa = 10.22VVRR538 pKa = 11.84ITVTAVNDD546 pKa = 3.78VPVTTPAAFTTGPGVAVTDD565 pKa = 3.95TVAWHH570 pKa = 6.63ASDD573 pKa = 4.78VDD575 pKa = 3.98VADD578 pKa = 4.25EE579 pKa = 4.33LTFTAAAPTAGTVTMGADD597 pKa = 3.52GQFTYY602 pKa = 10.19TPRR605 pKa = 11.84PGFSGTDD612 pKa = 3.04TFVFTVTDD620 pKa = 3.64TAGATATGVVSVTVANQQPVAYY642 pKa = 9.58DD643 pKa = 3.19DD644 pKa = 5.28GYY646 pKa = 11.47QVTPGATLTVDD657 pKa = 3.41AAHH660 pKa = 6.97GLLANDD666 pKa = 4.32VDD668 pKa = 4.53PGDD671 pKa = 4.37PLTVTSVTQPTHH683 pKa = 4.79GTVTVAGDD691 pKa = 3.47GSFTYY696 pKa = 10.21TPTAGFSGDD705 pKa = 3.2DD706 pKa = 3.29WFSYY710 pKa = 10.33TITDD714 pKa = 3.68SAGATATATVFFGVAPASNLPPIAPDD740 pKa = 2.96WTLTTEE746 pKa = 4.17ADD748 pKa = 3.14VDD750 pKa = 4.13VTFDD754 pKa = 3.78PLAGVTDD761 pKa = 4.33PNGDD765 pKa = 3.92VLWVRR770 pKa = 11.84DD771 pKa = 4.11GEE773 pKa = 4.43QPAHH777 pKa = 6.65GFVQQYY783 pKa = 9.57TNGTIAYY790 pKa = 8.86VPDD793 pKa = 3.62GTFVGTDD800 pKa = 2.89GFTYY804 pKa = 10.05EE805 pKa = 4.21VHH807 pKa = 7.5DD808 pKa = 3.96GTNVVTVRR816 pKa = 11.84VTVITSPSTAPVATGDD832 pKa = 3.83EE833 pKa = 4.31VSMPEE838 pKa = 4.21GGTVTIDD845 pKa = 3.24PLANDD850 pKa = 4.21TFIGTPTVEE859 pKa = 5.05LISTAQYY866 pKa = 8.99GTVTVDD872 pKa = 2.77AATGKK877 pKa = 8.53LTYY880 pKa = 10.44VATAGTNALGDD891 pKa = 3.71SFAYY895 pKa = 10.29QITDD899 pKa = 2.9EE900 pKa = 4.2RR901 pKa = 11.84GRR903 pKa = 11.84WSVASVDD910 pKa = 3.34ITFTAGNRR918 pKa = 11.84PPVATDD924 pKa = 3.85DD925 pKa = 3.96VAATPIGTPVSIDD938 pKa = 3.44VLANDD943 pKa = 4.08YY944 pKa = 11.44DD945 pKa = 4.85RR946 pKa = 11.84DD947 pKa = 3.97GTTLTVSVVSQPEE960 pKa = 3.88FGGTAIARR968 pKa = 11.84KK969 pKa = 10.07DD970 pKa = 3.36GTIDD974 pKa = 3.49FTPTKK979 pKa = 9.41GYY981 pKa = 11.04VGMASFSYY989 pKa = 10.42EE990 pKa = 3.95VTDD993 pKa = 3.51EE994 pKa = 4.24SGAVARR1000 pKa = 11.84ATVTVTVYY1008 pKa = 11.11DD1009 pKa = 4.18PTPPDD1014 pKa = 3.93APVKK1018 pKa = 10.51SYY1020 pKa = 9.76TVGKK1024 pKa = 10.42GRR1026 pKa = 11.84TLTVSLGDD1034 pKa = 3.65GVLAGSGYY1042 pKa = 10.18QGWLTGLEE1050 pKa = 4.08TGPAHH1055 pKa = 6.33GVLRR1059 pKa = 11.84LAEE1062 pKa = 5.09DD1063 pKa = 4.02GSFTYY1068 pKa = 9.86TPRR1071 pKa = 11.84EE1072 pKa = 4.09GFVGTDD1078 pKa = 2.89SFVFYY1083 pKa = 10.99VHH1085 pKa = 6.24TRR1087 pKa = 11.84EE1088 pKa = 4.23QGDD1091 pKa = 4.26GPFTATITVTEE1102 pKa = 5.25DD1103 pKa = 4.04GPGTPPNDD1111 pKa = 3.4AVAVALSSSDD1121 pKa = 3.28TAYY1124 pKa = 8.34GTCGGDD1130 pKa = 2.8WFAGRR1135 pKa = 11.84DD1136 pKa = 3.95DD1137 pKa = 5.0PASTWPTCLFLGRR1150 pKa = 11.84PEE1152 pKa = 4.31DD1153 pKa = 3.65VV1154 pKa = 3.47
Molecular weight: 117.99 kDa
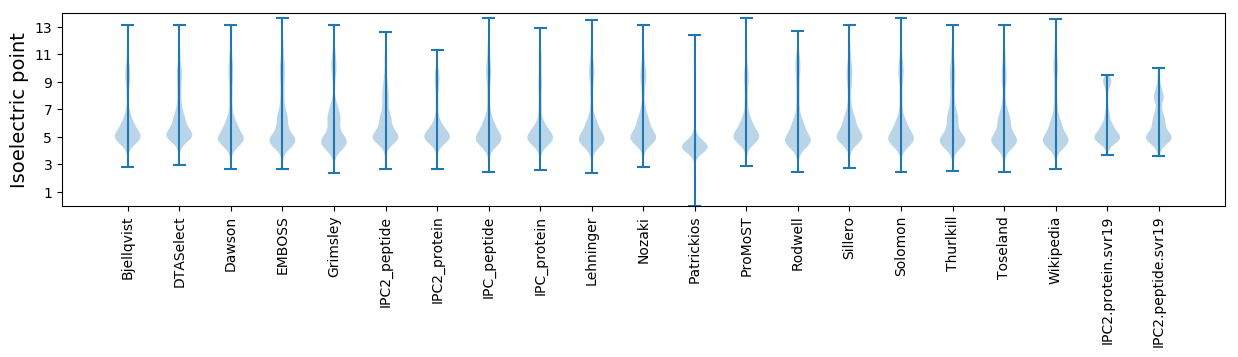
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E8Q1F6|A0A1E8Q1F6_9MYCO Molybdopterin molybdenumtransferase OS=Mycobacterium grossiae OX=1552759 GN=BEL07_20890 PE=3 SV=1
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVSSRR37 pKa = 11.84RR38 pKa = 11.84SKK40 pKa = 10.72GRR42 pKa = 11.84RR43 pKa = 11.84SLTAA47 pKa = 3.9
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVSSRR37 pKa = 11.84RR38 pKa = 11.84SKK40 pKa = 10.72GRR42 pKa = 11.84RR43 pKa = 11.84SLTAA47 pKa = 3.9
Molecular weight: 5.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1755592 |
29 |
4278 |
316.8 |
33.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.753 ± 0.047 | 0.754 ± 0.009 |
6.683 ± 0.029 | 5.017 ± 0.029 |
2.966 ± 0.016 | 8.991 ± 0.034 |
2.207 ± 0.017 | 3.825 ± 0.019 |
1.895 ± 0.023 | 9.846 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.916 ± 0.015 | 1.973 ± 0.018 |
5.806 ± 0.029 | 2.712 ± 0.017 |
7.52 ± 0.041 | 5.092 ± 0.024 |
6.311 ± 0.03 | 9.168 ± 0.036 |
1.493 ± 0.014 | 2.074 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
