
Podoviridae sp. ctdc61
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; unclassified Podoviridae
Average proteome isoelectric point is 5.47
Get precalculated fractions of proteins
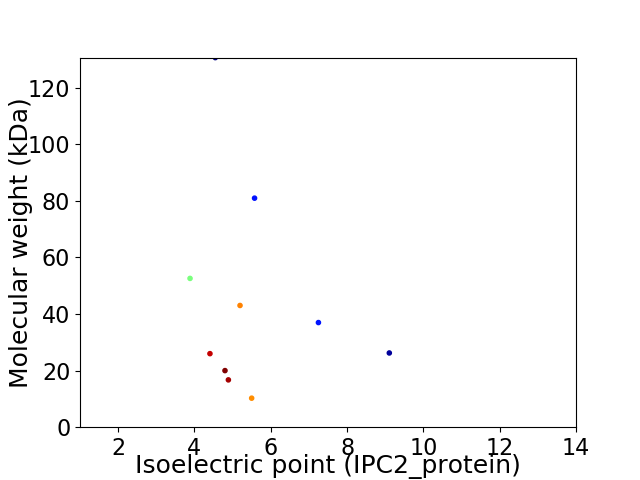
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A385E2G4|A0A385E2G4_9CAUD YvrJ family protein OS=Podoviridae sp. ctdc61 OX=2675449 PE=4 SV=1
MM1 pKa = 7.44TYY3 pKa = 10.27DD4 pKa = 3.78YY5 pKa = 11.14KK6 pKa = 11.42NKK8 pKa = 10.15AVNVRR13 pKa = 11.84QLNNYY18 pKa = 8.25MYY20 pKa = 11.1AKK22 pKa = 7.97TLSMFEE28 pKa = 4.17YY29 pKa = 10.29QGLPPSIPVRR39 pKa = 11.84EE40 pKa = 4.35LEE42 pKa = 4.11RR43 pKa = 11.84LLQIGGYY50 pKa = 10.61AFITEE55 pKa = 4.24APNGKK60 pKa = 9.9LYY62 pKa = 10.84AFHH65 pKa = 7.17GGLGGVPCPYY75 pKa = 10.66GNPTEE80 pKa = 4.43IVIANPALNFYY91 pKa = 8.42KK92 pKa = 10.11TLKK95 pKa = 10.57LKK97 pKa = 10.02TDD99 pKa = 3.49GVLIRR104 pKa = 11.84SDD106 pKa = 4.54DD107 pKa = 3.62MALGLDD113 pKa = 3.49PLFYY117 pKa = 10.37KK118 pKa = 10.81CNMLLAEE125 pKa = 4.67NDD127 pKa = 3.14INMILHH133 pKa = 6.91GYY135 pKa = 6.18STRR138 pKa = 11.84MQKK141 pKa = 10.58LISASDD147 pKa = 3.4DD148 pKa = 3.44KK149 pKa = 11.28TRR151 pKa = 11.84EE152 pKa = 4.05SADD155 pKa = 3.47SYY157 pKa = 10.57LKK159 pKa = 10.62KK160 pKa = 10.99VIDD163 pKa = 4.07GEE165 pKa = 4.36VSIVAEE171 pKa = 3.89NALFDD176 pKa = 3.71GVKK179 pKa = 9.62IHH181 pKa = 5.96GTSGDD186 pKa = 3.96SVGVTGMVEE195 pKa = 3.19YY196 pKa = 10.08HH197 pKa = 6.18QYY199 pKa = 10.74VKK201 pKa = 10.9SLMLSEE207 pKa = 5.02IGLSTSFNMKK217 pKa = 9.31KK218 pKa = 10.34EE219 pKa = 4.01RR220 pKa = 11.84MITAEE225 pKa = 4.46VDD227 pKa = 3.42QQDD230 pKa = 4.07DD231 pKa = 4.02STFPFVYY238 pKa = 11.02NMMKK242 pKa = 10.25CRR244 pKa = 11.84LHH246 pKa = 7.38GMEE249 pKa = 5.93RR250 pKa = 11.84INEE253 pKa = 4.13MYY255 pKa = 8.52GTSIEE260 pKa = 4.07VDD262 pKa = 4.66FGSVWNLKK270 pKa = 9.73NKK272 pKa = 9.96QLVDD276 pKa = 4.52DD277 pKa = 5.31IIGNNEE283 pKa = 4.42DD284 pKa = 3.9EE285 pKa = 4.99LSTIDD290 pKa = 4.82GEE292 pKa = 4.36IDD294 pKa = 3.47NEE296 pKa = 4.72LVQLDD301 pKa = 4.27NDD303 pKa = 4.1DD304 pKa = 3.95TTDD307 pKa = 3.81DD308 pKa = 4.12DD309 pKa = 5.9LIDD312 pKa = 4.53RR313 pKa = 11.84EE314 pKa = 4.19LAEE317 pKa = 5.44LEE319 pKa = 4.43TLSSALDD326 pKa = 3.53KK327 pKa = 10.84PLEE330 pKa = 4.58DD331 pKa = 4.69EE332 pKa = 5.59DD333 pKa = 5.07VEE335 pKa = 4.24LDD337 pKa = 3.53KK338 pKa = 11.68LIDD341 pKa = 3.75EE342 pKa = 4.84VVDD345 pKa = 3.59EE346 pKa = 4.75TVDD349 pKa = 3.36EE350 pKa = 4.28EE351 pKa = 4.69LFEE354 pKa = 5.3KK355 pKa = 10.73EE356 pKa = 4.02IDD358 pKa = 4.0DD359 pKa = 4.93GLAEE363 pKa = 5.03LDD365 pKa = 3.55PTKK368 pKa = 10.8TEE370 pKa = 4.7DD371 pKa = 4.21EE372 pKa = 4.39DD373 pKa = 5.62DD374 pKa = 3.92EE375 pKa = 4.98PEE377 pKa = 4.8PEE379 pKa = 4.13TSVTDD384 pKa = 3.49IEE386 pKa = 4.35DD387 pKa = 3.8TEE389 pKa = 4.64RR390 pKa = 11.84DD391 pKa = 3.5IPEE394 pKa = 4.27TEE396 pKa = 4.36LDD398 pKa = 4.07KK399 pKa = 11.65DD400 pKa = 5.31DD401 pKa = 6.35DD402 pKa = 4.47DD403 pKa = 4.11EE404 pKa = 4.68QNAVVEE410 pKa = 5.12DD411 pKa = 3.79IVDD414 pKa = 4.2DD415 pKa = 4.17SGTDD419 pKa = 3.38NEE421 pKa = 4.33QSQEE425 pKa = 3.99EE426 pKa = 4.72VVSVEE431 pKa = 4.64VIDD434 pKa = 3.95TVIDD438 pKa = 3.5VLEE441 pKa = 4.49SIKK444 pKa = 10.22TSTNDD449 pKa = 3.3EE450 pKa = 4.08IEE452 pKa = 4.79EE453 pKa = 4.38ISDD456 pKa = 3.64TLNTSKK462 pKa = 11.19DD463 pKa = 3.2KK464 pKa = 11.6DD465 pKa = 3.73NASTT469 pKa = 3.54
MM1 pKa = 7.44TYY3 pKa = 10.27DD4 pKa = 3.78YY5 pKa = 11.14KK6 pKa = 11.42NKK8 pKa = 10.15AVNVRR13 pKa = 11.84QLNNYY18 pKa = 8.25MYY20 pKa = 11.1AKK22 pKa = 7.97TLSMFEE28 pKa = 4.17YY29 pKa = 10.29QGLPPSIPVRR39 pKa = 11.84EE40 pKa = 4.35LEE42 pKa = 4.11RR43 pKa = 11.84LLQIGGYY50 pKa = 10.61AFITEE55 pKa = 4.24APNGKK60 pKa = 9.9LYY62 pKa = 10.84AFHH65 pKa = 7.17GGLGGVPCPYY75 pKa = 10.66GNPTEE80 pKa = 4.43IVIANPALNFYY91 pKa = 8.42KK92 pKa = 10.11TLKK95 pKa = 10.57LKK97 pKa = 10.02TDD99 pKa = 3.49GVLIRR104 pKa = 11.84SDD106 pKa = 4.54DD107 pKa = 3.62MALGLDD113 pKa = 3.49PLFYY117 pKa = 10.37KK118 pKa = 10.81CNMLLAEE125 pKa = 4.67NDD127 pKa = 3.14INMILHH133 pKa = 6.91GYY135 pKa = 6.18STRR138 pKa = 11.84MQKK141 pKa = 10.58LISASDD147 pKa = 3.4DD148 pKa = 3.44KK149 pKa = 11.28TRR151 pKa = 11.84EE152 pKa = 4.05SADD155 pKa = 3.47SYY157 pKa = 10.57LKK159 pKa = 10.62KK160 pKa = 10.99VIDD163 pKa = 4.07GEE165 pKa = 4.36VSIVAEE171 pKa = 3.89NALFDD176 pKa = 3.71GVKK179 pKa = 9.62IHH181 pKa = 5.96GTSGDD186 pKa = 3.96SVGVTGMVEE195 pKa = 3.19YY196 pKa = 10.08HH197 pKa = 6.18QYY199 pKa = 10.74VKK201 pKa = 10.9SLMLSEE207 pKa = 5.02IGLSTSFNMKK217 pKa = 9.31KK218 pKa = 10.34EE219 pKa = 4.01RR220 pKa = 11.84MITAEE225 pKa = 4.46VDD227 pKa = 3.42QQDD230 pKa = 4.07DD231 pKa = 4.02STFPFVYY238 pKa = 11.02NMMKK242 pKa = 10.25CRR244 pKa = 11.84LHH246 pKa = 7.38GMEE249 pKa = 5.93RR250 pKa = 11.84INEE253 pKa = 4.13MYY255 pKa = 8.52GTSIEE260 pKa = 4.07VDD262 pKa = 4.66FGSVWNLKK270 pKa = 9.73NKK272 pKa = 9.96QLVDD276 pKa = 4.52DD277 pKa = 5.31IIGNNEE283 pKa = 4.42DD284 pKa = 3.9EE285 pKa = 4.99LSTIDD290 pKa = 4.82GEE292 pKa = 4.36IDD294 pKa = 3.47NEE296 pKa = 4.72LVQLDD301 pKa = 4.27NDD303 pKa = 4.1DD304 pKa = 3.95TTDD307 pKa = 3.81DD308 pKa = 4.12DD309 pKa = 5.9LIDD312 pKa = 4.53RR313 pKa = 11.84EE314 pKa = 4.19LAEE317 pKa = 5.44LEE319 pKa = 4.43TLSSALDD326 pKa = 3.53KK327 pKa = 10.84PLEE330 pKa = 4.58DD331 pKa = 4.69EE332 pKa = 5.59DD333 pKa = 5.07VEE335 pKa = 4.24LDD337 pKa = 3.53KK338 pKa = 11.68LIDD341 pKa = 3.75EE342 pKa = 4.84VVDD345 pKa = 3.59EE346 pKa = 4.75TVDD349 pKa = 3.36EE350 pKa = 4.28EE351 pKa = 4.69LFEE354 pKa = 5.3KK355 pKa = 10.73EE356 pKa = 4.02IDD358 pKa = 4.0DD359 pKa = 4.93GLAEE363 pKa = 5.03LDD365 pKa = 3.55PTKK368 pKa = 10.8TEE370 pKa = 4.7DD371 pKa = 4.21EE372 pKa = 4.39DD373 pKa = 5.62DD374 pKa = 3.92EE375 pKa = 4.98PEE377 pKa = 4.8PEE379 pKa = 4.13TSVTDD384 pKa = 3.49IEE386 pKa = 4.35DD387 pKa = 3.8TEE389 pKa = 4.64RR390 pKa = 11.84DD391 pKa = 3.5IPEE394 pKa = 4.27TEE396 pKa = 4.36LDD398 pKa = 4.07KK399 pKa = 11.65DD400 pKa = 5.31DD401 pKa = 6.35DD402 pKa = 4.47DD403 pKa = 4.11EE404 pKa = 4.68QNAVVEE410 pKa = 5.12DD411 pKa = 3.79IVDD414 pKa = 4.2DD415 pKa = 4.17SGTDD419 pKa = 3.38NEE421 pKa = 4.33QSQEE425 pKa = 3.99EE426 pKa = 4.72VVSVEE431 pKa = 4.64VIDD434 pKa = 3.95TVIDD438 pKa = 3.5VLEE441 pKa = 4.49SIKK444 pKa = 10.22TSTNDD449 pKa = 3.3EE450 pKa = 4.08IEE452 pKa = 4.79EE453 pKa = 4.38ISDD456 pKa = 3.64TLNTSKK462 pKa = 11.19DD463 pKa = 3.2KK464 pKa = 11.6DD465 pKa = 3.73NASTT469 pKa = 3.54
Molecular weight: 52.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A385E503|A0A385E503_9CAUD Uncharacterized protein OS=Podoviridae sp. ctdc61 OX=2675449 PE=4 SV=1
MM1 pKa = 7.87ALPAILIGVSRR12 pKa = 11.84LALGTQRR19 pKa = 11.84FRR21 pKa = 11.84SAATVTTGIGVRR33 pKa = 11.84SNVVRR38 pKa = 11.84VPTIAAQQQQNKK50 pKa = 9.03KK51 pKa = 9.38RR52 pKa = 11.84LRR54 pKa = 11.84EE55 pKa = 4.02LASSLASKK63 pKa = 10.3ANKK66 pKa = 9.16RR67 pKa = 11.84VRR69 pKa = 11.84RR70 pKa = 11.84LEE72 pKa = 4.02KK73 pKa = 10.94NGFTDD78 pKa = 3.42QSAYY82 pKa = 8.46RR83 pKa = 11.84TYY85 pKa = 10.97VEE87 pKa = 4.24SGGKK91 pKa = 10.09FSVKK95 pKa = 9.8GKK97 pKa = 9.42NEE99 pKa = 3.86KK100 pKa = 10.32EE101 pKa = 3.63ILAEE105 pKa = 3.89LEE107 pKa = 4.07RR108 pKa = 11.84VKK110 pKa = 10.91KK111 pKa = 10.55FLGDD115 pKa = 3.45EE116 pKa = 4.21SSTVSGINKK125 pKa = 8.69QAEE128 pKa = 4.42SVAKK132 pKa = 10.24RR133 pKa = 11.84LNIKK137 pKa = 10.35YY138 pKa = 8.36KK139 pKa = 8.14TFKK142 pKa = 10.31EE143 pKa = 4.0LRR145 pKa = 11.84EE146 pKa = 3.91KK147 pKa = 10.66NQAIFKK153 pKa = 8.93LLPKK157 pKa = 8.74ITEE160 pKa = 4.01YY161 pKa = 10.89MKK163 pKa = 10.38MSSNTYY169 pKa = 8.74TEE171 pKa = 4.06YY172 pKa = 11.28DD173 pKa = 3.31SDD175 pKa = 3.95QVIDD179 pKa = 4.02EE180 pKa = 4.32VTDD183 pKa = 3.72YY184 pKa = 11.22VDD186 pKa = 4.25EE187 pKa = 4.73VMAQLKK193 pKa = 10.63DD194 pKa = 3.5GEE196 pKa = 4.54TLDD199 pKa = 4.31IEE201 pKa = 4.82QAIQDD206 pKa = 3.46IEE208 pKa = 3.99QRR210 pKa = 11.84IIKK213 pKa = 9.68LDD215 pKa = 3.44KK216 pKa = 10.66KK217 pKa = 10.38SISVKK222 pKa = 10.17RR223 pKa = 11.84SNIPSFYY230 pKa = 10.74KK231 pKa = 10.51PP232 pKa = 3.21
MM1 pKa = 7.87ALPAILIGVSRR12 pKa = 11.84LALGTQRR19 pKa = 11.84FRR21 pKa = 11.84SAATVTTGIGVRR33 pKa = 11.84SNVVRR38 pKa = 11.84VPTIAAQQQQNKK50 pKa = 9.03KK51 pKa = 9.38RR52 pKa = 11.84LRR54 pKa = 11.84EE55 pKa = 4.02LASSLASKK63 pKa = 10.3ANKK66 pKa = 9.16RR67 pKa = 11.84VRR69 pKa = 11.84RR70 pKa = 11.84LEE72 pKa = 4.02KK73 pKa = 10.94NGFTDD78 pKa = 3.42QSAYY82 pKa = 8.46RR83 pKa = 11.84TYY85 pKa = 10.97VEE87 pKa = 4.24SGGKK91 pKa = 10.09FSVKK95 pKa = 9.8GKK97 pKa = 9.42NEE99 pKa = 3.86KK100 pKa = 10.32EE101 pKa = 3.63ILAEE105 pKa = 3.89LEE107 pKa = 4.07RR108 pKa = 11.84VKK110 pKa = 10.91KK111 pKa = 10.55FLGDD115 pKa = 3.45EE116 pKa = 4.21SSTVSGINKK125 pKa = 8.69QAEE128 pKa = 4.42SVAKK132 pKa = 10.24RR133 pKa = 11.84LNIKK137 pKa = 10.35YY138 pKa = 8.36KK139 pKa = 8.14TFKK142 pKa = 10.31EE143 pKa = 4.0LRR145 pKa = 11.84EE146 pKa = 3.91KK147 pKa = 10.66NQAIFKK153 pKa = 8.93LLPKK157 pKa = 8.74ITEE160 pKa = 4.01YY161 pKa = 10.89MKK163 pKa = 10.38MSSNTYY169 pKa = 8.74TEE171 pKa = 4.06YY172 pKa = 11.28DD173 pKa = 3.31SDD175 pKa = 3.95QVIDD179 pKa = 4.02EE180 pKa = 4.32VTDD183 pKa = 3.72YY184 pKa = 11.22VDD186 pKa = 4.25EE187 pKa = 4.73VMAQLKK193 pKa = 10.63DD194 pKa = 3.5GEE196 pKa = 4.54TLDD199 pKa = 4.31IEE201 pKa = 4.82QAIQDD206 pKa = 3.46IEE208 pKa = 3.99QRR210 pKa = 11.84IIKK213 pKa = 9.68LDD215 pKa = 3.44KK216 pKa = 10.66KK217 pKa = 10.38SISVKK222 pKa = 10.17RR223 pKa = 11.84SNIPSFYY230 pKa = 10.74KK231 pKa = 10.51PP232 pKa = 3.21
Molecular weight: 26.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3940 |
90 |
1186 |
394.0 |
44.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.924 ± 0.265 | 0.635 ± 0.297 |
7.766 ± 0.728 | 6.244 ± 0.74 |
4.01 ± 0.333 | 5.914 ± 0.535 |
1.168 ± 0.221 | 7.766 ± 0.533 |
6.777 ± 0.487 | 7.919 ± 0.439 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.234 ± 0.35 | 6.777 ± 0.419 |
3.756 ± 0.762 | 2.157 ± 0.339 |
3.325 ± 0.365 | 6.777 ± 0.328 |
7.893 ± 0.407 | 8.02 ± 0.69 |
0.939 ± 0.134 | 5.0 ± 0.49 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
