
Kiritimatiellaeota bacterium S-5007
Taxonomy: cellular organisms; Bacteria; PVC group; Kiritimatiellaeota; unclassified Kiritimatiellaeota
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
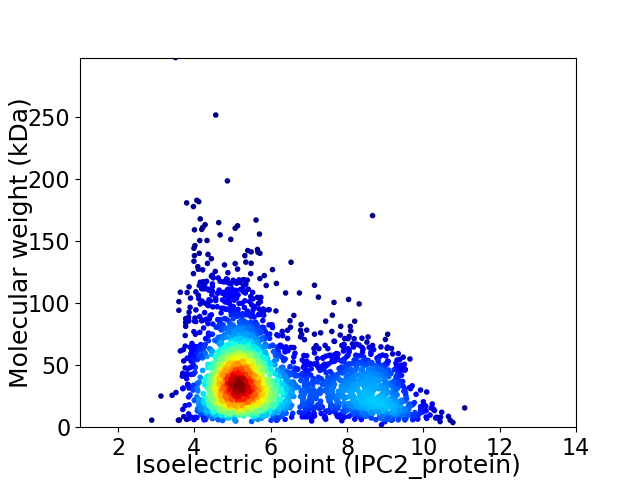
Virtual 2D-PAGE plot for 3062 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P1M4Q9|A0A6P1M4Q9_9BACT Uncharacterized protein OS=Kiritimatiellaeota bacterium S-5007 OX=2697043 GN=GT409_04990 PE=3 SV=1
MM1 pKa = 7.42TEE3 pKa = 4.18SKK5 pKa = 10.91PMVKK9 pKa = 10.02KK10 pKa = 10.9GILLSMLLAAICHH23 pKa = 6.3SDD25 pKa = 3.67PVDD28 pKa = 4.04GLDD31 pKa = 3.8GMAGSSTMDD40 pKa = 4.06EE41 pKa = 4.08IEE43 pKa = 4.14DD44 pKa = 3.68WFVVTGSVQSVTNGYY59 pKa = 9.15GAVGMISAAGSNSLWTAEE77 pKa = 4.19YY78 pKa = 10.57VSSDD82 pKa = 3.51VLQTNAVYY90 pKa = 10.68GLAVRR95 pKa = 11.84AGNWSSNPGSGTVFAIAIGYY115 pKa = 8.89YY116 pKa = 9.94DD117 pKa = 4.25SEE119 pKa = 4.16WHH121 pKa = 6.38EE122 pKa = 4.29LTNKK126 pKa = 8.89TFTGGTAGEE135 pKa = 4.12ITPTINGSYY144 pKa = 7.36EE145 pKa = 3.99TLVFTNEE152 pKa = 3.6LAVFANVAVRR162 pKa = 11.84VGRR165 pKa = 11.84TGTSGSWGGFDD176 pKa = 3.54TVSLTILQSDD186 pKa = 3.71SDD188 pKa = 4.43GDD190 pKa = 3.95GLPDD194 pKa = 3.09RR195 pKa = 11.84WEE197 pKa = 3.81FDD199 pKa = 3.33YY200 pKa = 11.56GLNPNDD206 pKa = 3.45GGTFNFDD213 pKa = 4.58DD214 pKa = 5.47GDD216 pKa = 4.49LGDD219 pKa = 5.24PDD221 pKa = 5.94DD222 pKa = 6.79DD223 pKa = 4.21GLQNHH228 pKa = 5.73QEE230 pKa = 4.06YY231 pKa = 11.02SLGSDD236 pKa = 3.52PLVDD240 pKa = 3.32EE241 pKa = 4.65WKK243 pKa = 10.61SRR245 pKa = 11.84PEE247 pKa = 3.64KK248 pKa = 10.56ARR250 pKa = 11.84LMVINAHH257 pKa = 6.98PDD259 pKa = 3.08DD260 pKa = 3.84EE261 pKa = 6.91AIFFGGAIPYY271 pKa = 6.95YY272 pKa = 8.06TQVRR276 pKa = 11.84QLPTIVISMTSGLVSEE292 pKa = 4.81TEE294 pKa = 3.43IHH296 pKa = 5.38EE297 pKa = 4.38AEE299 pKa = 4.31FRR301 pKa = 11.84SACWAYY307 pKa = 10.44GLRR310 pKa = 11.84NQPVFARR317 pKa = 11.84FYY319 pKa = 9.31NNVWNASLDD328 pKa = 4.07EE329 pKa = 4.46TWDD332 pKa = 3.15TWADD336 pKa = 3.55GVLDD340 pKa = 4.52GDD342 pKa = 6.08DD343 pKa = 3.46ITEE346 pKa = 4.37GQALAAEE353 pKa = 4.4TLTYY357 pKa = 9.53WIRR360 pKa = 11.84RR361 pKa = 11.84YY362 pKa = 10.15QPDD365 pKa = 3.5VVVTHH370 pKa = 6.83DD371 pKa = 3.71LEE373 pKa = 5.77GEE375 pKa = 4.32YY376 pKa = 10.98GHH378 pKa = 6.79SDD380 pKa = 3.98HH381 pKa = 7.09IATAITATNAVQLAADD397 pKa = 3.78TNYY400 pKa = 10.06VDD402 pKa = 6.09GLQPWQVKK410 pKa = 9.82KK411 pKa = 10.67LYY413 pKa = 10.89LHH415 pKa = 6.98MYY417 pKa = 6.8PTNQLFHH424 pKa = 7.44DD425 pKa = 4.91FWQDD429 pKa = 2.88VSIDD433 pKa = 3.68TNADD437 pKa = 2.75ATADD441 pKa = 3.74ITPMDD446 pKa = 4.25AADD449 pKa = 3.61IGLAFHH455 pKa = 7.22VYY457 pKa = 10.12QGTPTNASTVYY468 pKa = 10.48AVNEE472 pKa = 4.03TSDD475 pKa = 4.5AGWEE479 pKa = 4.16PYY481 pKa = 9.71PSEE484 pKa = 3.37WWGLVSSTVGADD496 pKa = 3.03TATSFTAPDD505 pKa = 3.8ADD507 pKa = 3.56NVEE510 pKa = 3.85ISYY513 pKa = 10.26VGWARR518 pKa = 11.84GDD520 pKa = 3.64FFEE523 pKa = 6.11HH524 pKa = 5.45ITVFADD530 pKa = 3.02SDD532 pKa = 4.21YY533 pKa = 11.73DD534 pKa = 3.63QLPDD538 pKa = 3.54DD539 pKa = 4.25WEE541 pKa = 4.3LAHH544 pKa = 6.8FPSLEE549 pKa = 4.04AADD552 pKa = 4.75PDD554 pKa = 4.48ADD556 pKa = 4.2PDD558 pKa = 4.97GDD560 pKa = 4.08GLDD563 pKa = 3.68NAAEE567 pKa = 4.46FGAGTNPALSDD578 pKa = 3.44TDD580 pKa = 3.52GDD582 pKa = 4.22SFNDD586 pKa = 3.57GLEE589 pKa = 4.36VINGGDD595 pKa = 3.91PLVSDD600 pKa = 4.76LWRR603 pKa = 11.84IDD605 pKa = 3.67YY606 pKa = 10.81IRR608 pKa = 11.84DD609 pKa = 3.15NGADD613 pKa = 3.52YY614 pKa = 11.42DD615 pKa = 4.46LYY617 pKa = 11.4SSNAVLDD624 pKa = 4.02LSIGQAAFAVSGGTAWLALQLEE646 pKa = 4.45KK647 pKa = 11.04SEE649 pKa = 6.36DD650 pKa = 3.99LITWTNAGDD659 pKa = 3.81TVIWSIPVNTNNAFYY674 pKa = 10.44RR675 pKa = 11.84VRR677 pKa = 11.84SSRR680 pKa = 3.29
MM1 pKa = 7.42TEE3 pKa = 4.18SKK5 pKa = 10.91PMVKK9 pKa = 10.02KK10 pKa = 10.9GILLSMLLAAICHH23 pKa = 6.3SDD25 pKa = 3.67PVDD28 pKa = 4.04GLDD31 pKa = 3.8GMAGSSTMDD40 pKa = 4.06EE41 pKa = 4.08IEE43 pKa = 4.14DD44 pKa = 3.68WFVVTGSVQSVTNGYY59 pKa = 9.15GAVGMISAAGSNSLWTAEE77 pKa = 4.19YY78 pKa = 10.57VSSDD82 pKa = 3.51VLQTNAVYY90 pKa = 10.68GLAVRR95 pKa = 11.84AGNWSSNPGSGTVFAIAIGYY115 pKa = 8.89YY116 pKa = 9.94DD117 pKa = 4.25SEE119 pKa = 4.16WHH121 pKa = 6.38EE122 pKa = 4.29LTNKK126 pKa = 8.89TFTGGTAGEE135 pKa = 4.12ITPTINGSYY144 pKa = 7.36EE145 pKa = 3.99TLVFTNEE152 pKa = 3.6LAVFANVAVRR162 pKa = 11.84VGRR165 pKa = 11.84TGTSGSWGGFDD176 pKa = 3.54TVSLTILQSDD186 pKa = 3.71SDD188 pKa = 4.43GDD190 pKa = 3.95GLPDD194 pKa = 3.09RR195 pKa = 11.84WEE197 pKa = 3.81FDD199 pKa = 3.33YY200 pKa = 11.56GLNPNDD206 pKa = 3.45GGTFNFDD213 pKa = 4.58DD214 pKa = 5.47GDD216 pKa = 4.49LGDD219 pKa = 5.24PDD221 pKa = 5.94DD222 pKa = 6.79DD223 pKa = 4.21GLQNHH228 pKa = 5.73QEE230 pKa = 4.06YY231 pKa = 11.02SLGSDD236 pKa = 3.52PLVDD240 pKa = 3.32EE241 pKa = 4.65WKK243 pKa = 10.61SRR245 pKa = 11.84PEE247 pKa = 3.64KK248 pKa = 10.56ARR250 pKa = 11.84LMVINAHH257 pKa = 6.98PDD259 pKa = 3.08DD260 pKa = 3.84EE261 pKa = 6.91AIFFGGAIPYY271 pKa = 6.95YY272 pKa = 8.06TQVRR276 pKa = 11.84QLPTIVISMTSGLVSEE292 pKa = 4.81TEE294 pKa = 3.43IHH296 pKa = 5.38EE297 pKa = 4.38AEE299 pKa = 4.31FRR301 pKa = 11.84SACWAYY307 pKa = 10.44GLRR310 pKa = 11.84NQPVFARR317 pKa = 11.84FYY319 pKa = 9.31NNVWNASLDD328 pKa = 4.07EE329 pKa = 4.46TWDD332 pKa = 3.15TWADD336 pKa = 3.55GVLDD340 pKa = 4.52GDD342 pKa = 6.08DD343 pKa = 3.46ITEE346 pKa = 4.37GQALAAEE353 pKa = 4.4TLTYY357 pKa = 9.53WIRR360 pKa = 11.84RR361 pKa = 11.84YY362 pKa = 10.15QPDD365 pKa = 3.5VVVTHH370 pKa = 6.83DD371 pKa = 3.71LEE373 pKa = 5.77GEE375 pKa = 4.32YY376 pKa = 10.98GHH378 pKa = 6.79SDD380 pKa = 3.98HH381 pKa = 7.09IATAITATNAVQLAADD397 pKa = 3.78TNYY400 pKa = 10.06VDD402 pKa = 6.09GLQPWQVKK410 pKa = 9.82KK411 pKa = 10.67LYY413 pKa = 10.89LHH415 pKa = 6.98MYY417 pKa = 6.8PTNQLFHH424 pKa = 7.44DD425 pKa = 4.91FWQDD429 pKa = 2.88VSIDD433 pKa = 3.68TNADD437 pKa = 2.75ATADD441 pKa = 3.74ITPMDD446 pKa = 4.25AADD449 pKa = 3.61IGLAFHH455 pKa = 7.22VYY457 pKa = 10.12QGTPTNASTVYY468 pKa = 10.48AVNEE472 pKa = 4.03TSDD475 pKa = 4.5AGWEE479 pKa = 4.16PYY481 pKa = 9.71PSEE484 pKa = 3.37WWGLVSSTVGADD496 pKa = 3.03TATSFTAPDD505 pKa = 3.8ADD507 pKa = 3.56NVEE510 pKa = 3.85ISYY513 pKa = 10.26VGWARR518 pKa = 11.84GDD520 pKa = 3.64FFEE523 pKa = 6.11HH524 pKa = 5.45ITVFADD530 pKa = 3.02SDD532 pKa = 4.21YY533 pKa = 11.73DD534 pKa = 3.63QLPDD538 pKa = 3.54DD539 pKa = 4.25WEE541 pKa = 4.3LAHH544 pKa = 6.8FPSLEE549 pKa = 4.04AADD552 pKa = 4.75PDD554 pKa = 4.48ADD556 pKa = 4.2PDD558 pKa = 4.97GDD560 pKa = 4.08GLDD563 pKa = 3.68NAAEE567 pKa = 4.46FGAGTNPALSDD578 pKa = 3.44TDD580 pKa = 3.52GDD582 pKa = 4.22SFNDD586 pKa = 3.57GLEE589 pKa = 4.36VINGGDD595 pKa = 3.91PLVSDD600 pKa = 4.76LWRR603 pKa = 11.84IDD605 pKa = 3.67YY606 pKa = 10.81IRR608 pKa = 11.84DD609 pKa = 3.15NGADD613 pKa = 3.52YY614 pKa = 11.42DD615 pKa = 4.46LYY617 pKa = 11.4SSNAVLDD624 pKa = 4.02LSIGQAAFAVSGGTAWLALQLEE646 pKa = 4.45KK647 pKa = 11.04SEE649 pKa = 6.36DD650 pKa = 3.99LITWTNAGDD659 pKa = 3.81TVIWSIPVNTNNAFYY674 pKa = 10.44RR675 pKa = 11.84VRR677 pKa = 11.84SSRR680 pKa = 3.29
Molecular weight: 73.83 kDa
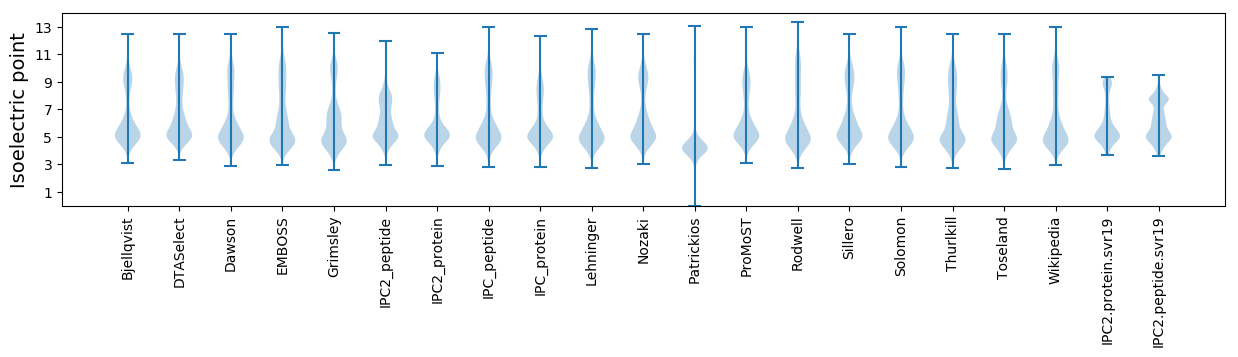
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P1M7S0|A0A6P1M7S0_9BACT Uncharacterized protein OS=Kiritimatiellaeota bacterium S-5007 OX=2697043 GN=GT409_15365 PE=3 SV=1
MM1 pKa = 7.07GTIKK5 pKa = 10.15KK6 pKa = 8.7WRR8 pKa = 11.84RR9 pKa = 11.84TKK11 pKa = 10.12MSNHH15 pKa = 5.85KK16 pKa = 9.51RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.42RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84ADD24 pKa = 3.32RR25 pKa = 11.84HH26 pKa = 5.42KK27 pKa = 11.04KK28 pKa = 9.18KK29 pKa = 10.86
MM1 pKa = 7.07GTIKK5 pKa = 10.15KK6 pKa = 8.7WRR8 pKa = 11.84RR9 pKa = 11.84TKK11 pKa = 10.12MSNHH15 pKa = 5.85KK16 pKa = 9.51RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.42RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84ADD24 pKa = 3.32RR25 pKa = 11.84HH26 pKa = 5.42KK27 pKa = 11.04KK28 pKa = 9.18KK29 pKa = 10.86
Molecular weight: 3.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1160248 |
17 |
2799 |
378.9 |
41.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.796 ± 0.044 | 1.317 ± 0.014 |
6.008 ± 0.034 | 6.509 ± 0.045 |
4.312 ± 0.025 | 7.92 ± 0.042 |
2.014 ± 0.019 | 5.684 ± 0.034 |
4.701 ± 0.043 | 9.402 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.503 ± 0.019 | 3.842 ± 0.028 |
4.662 ± 0.033 | 3.473 ± 0.021 |
5.464 ± 0.039 | 6.227 ± 0.042 |
5.507 ± 0.036 | 7.017 ± 0.036 |
1.56 ± 0.018 | 3.084 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
