
Prunus necrotic ringspot virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Ilarvirus
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
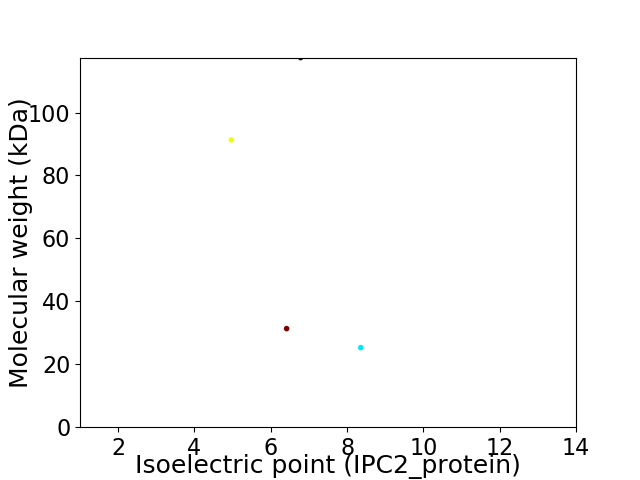
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q91NQ1|Q91NQ1_9BROM ATP-dependent helicase OS=Prunus necrotic ringspot virus OX=37733 PE=3 SV=1
MM1 pKa = 7.48NPLNQLLSHH10 pKa = 6.66GCSAISSALHH20 pKa = 6.39RR21 pKa = 11.84DD22 pKa = 3.17VAYY25 pKa = 8.84TQISDD30 pKa = 3.66EE31 pKa = 4.62CFDD34 pKa = 3.55SHH36 pKa = 6.35FTQISTEE43 pKa = 4.02YY44 pKa = 10.96SLTTSVRR51 pKa = 11.84KK52 pKa = 9.92LFEE55 pKa = 5.94AYY57 pKa = 8.83MVTMFLKK64 pKa = 10.5KK65 pKa = 9.59HH66 pKa = 5.03MSYY69 pKa = 10.78VFEE72 pKa = 4.88DD73 pKa = 4.92FIGSEE78 pKa = 4.05MGPYY82 pKa = 9.52VQTRR86 pKa = 11.84WRR88 pKa = 11.84IVFDD92 pKa = 3.91LDD94 pKa = 3.65EE95 pKa = 4.6VEE97 pKa = 4.48EE98 pKa = 4.66EE99 pKa = 4.42YY100 pKa = 11.07VYY102 pKa = 11.13KK103 pKa = 10.34PDD105 pKa = 3.98FYY107 pKa = 11.13HH108 pKa = 6.87YY109 pKa = 10.78DD110 pKa = 3.39VYY112 pKa = 10.99DD113 pKa = 3.64YY114 pKa = 11.71EE115 pKa = 4.46MGVDD119 pKa = 3.91IPASIMDD126 pKa = 6.05DD127 pKa = 3.3IPFCEE132 pKa = 4.47DD133 pKa = 3.44AEE135 pKa = 4.35AVTCRR140 pKa = 11.84NCGSFDD146 pKa = 3.52EE147 pKa = 4.86MEE149 pKa = 4.39TVYY152 pKa = 10.58QAATSSDD159 pKa = 3.16EE160 pKa = 4.13SRR162 pKa = 11.84ANVCDD167 pKa = 3.09VDD169 pKa = 4.8APHH172 pKa = 7.23DD173 pKa = 4.1TIKK176 pKa = 10.64WSDD179 pKa = 3.83EE180 pKa = 3.67PDD182 pKa = 3.45LTVDD186 pKa = 5.1DD187 pKa = 4.77PSKK190 pKa = 11.08LHH192 pKa = 7.11DD193 pKa = 5.38LDD195 pKa = 6.56DD196 pKa = 4.87EE197 pKa = 4.87VPQLQLEE204 pKa = 4.57TVGRR208 pKa = 11.84VVQSVEE214 pKa = 3.98PLVPDD219 pKa = 3.45INSFEE224 pKa = 4.32TNQLHH229 pKa = 5.98LVKK232 pKa = 10.4EE233 pKa = 4.43VVLDD237 pKa = 3.66RR238 pKa = 11.84FEE240 pKa = 6.52SILEE244 pKa = 3.99CHH246 pKa = 6.48EE247 pKa = 4.31KK248 pKa = 10.75VLARR252 pKa = 11.84NDD254 pKa = 3.44WKK256 pKa = 11.04SLAMYY261 pKa = 9.95VPTGEE266 pKa = 4.32IPSVSDD272 pKa = 3.06ILEE275 pKa = 4.18IPVKK279 pKa = 9.4HH280 pKa = 5.49TQPEE284 pKa = 4.95FIQMAIDD291 pKa = 3.56EE292 pKa = 4.74MLPGVSDD299 pKa = 3.64FDD301 pKa = 4.42DD302 pKa = 4.65RR303 pKa = 11.84FFQEE307 pKa = 3.89IVEE310 pKa = 4.3TSDD313 pKa = 2.94INLEE317 pKa = 3.73IDD319 pKa = 3.42RR320 pKa = 11.84ASIDD324 pKa = 3.21MSVFNDD330 pKa = 3.32WTPKK334 pKa = 9.87SGRR337 pKa = 11.84LNGLFQTGNISKK349 pKa = 10.21RR350 pKa = 11.84VPTFRR355 pKa = 11.84EE356 pKa = 3.29ASLAIKK362 pKa = 9.92KK363 pKa = 10.38RR364 pKa = 11.84NLNVPDD370 pKa = 4.15LQQVLHH376 pKa = 7.06EE377 pKa = 4.86DD378 pKa = 3.42DD379 pKa = 3.73EE380 pKa = 5.08ARR382 pKa = 11.84FIANKK387 pKa = 10.04FINTVIDD394 pKa = 4.11PNKK397 pKa = 9.5LAQFPGYY404 pKa = 9.77ISEE407 pKa = 4.62GEE409 pKa = 3.54MGYY412 pKa = 10.49FNKK415 pKa = 10.35YY416 pKa = 10.32LSGKK420 pKa = 6.74TVPDD424 pKa = 3.82DD425 pKa = 4.67AFVDD429 pKa = 4.29PCALVSMDD437 pKa = 4.87KK438 pKa = 9.87YY439 pKa = 10.69RR440 pKa = 11.84HH441 pKa = 5.51MIKK444 pKa = 8.45TQLKK448 pKa = 8.28PVEE451 pKa = 4.32DD452 pKa = 3.76TSQLFEE458 pKa = 4.66RR459 pKa = 11.84PLAATITYY467 pKa = 9.23HH468 pKa = 7.77DD469 pKa = 4.3KK470 pKa = 11.19GKK472 pKa = 10.97VMSTSPIFLMMCNRR486 pKa = 11.84LLLCLNDD493 pKa = 4.74KK494 pKa = 10.8ISIPSGKK501 pKa = 8.93HH502 pKa = 4.4HH503 pKa = 6.45QLFSLDD509 pKa = 3.38PFAFEE514 pKa = 3.71MTKK517 pKa = 10.08EE518 pKa = 4.05FKK520 pKa = 10.65EE521 pKa = 3.75IDD523 pKa = 3.38FSKK526 pKa = 10.51FDD528 pKa = 3.39KK529 pKa = 10.68SQQRR533 pKa = 11.84LHH535 pKa = 7.41HH536 pKa = 7.01LIQFHH541 pKa = 6.82IFTALGAPKK550 pKa = 10.28DD551 pKa = 3.96FLDD554 pKa = 3.35MWFASHH560 pKa = 7.23EE561 pKa = 4.05ISHH564 pKa = 6.59IRR566 pKa = 11.84DD567 pKa = 3.92GPCGIGFSVNYY578 pKa = 8.16QRR580 pKa = 11.84RR581 pKa = 11.84TGDD584 pKa = 2.69ACTYY588 pKa = 10.06LGNTIITLSALAYY601 pKa = 9.8MYY603 pKa = 10.97DD604 pKa = 3.7LLDD607 pKa = 4.26PNVTFVIASGDD618 pKa = 3.52DD619 pKa = 3.56SLIGSIKK626 pKa = 10.23PLDD629 pKa = 3.61RR630 pKa = 11.84SEE632 pKa = 4.05EE633 pKa = 4.18FKK635 pKa = 11.03FVTLFNFEE643 pKa = 4.35AKK645 pKa = 10.1FPHH648 pKa = 5.94NQPFVCSKK656 pKa = 9.39FLCLLPTVSGGKK668 pKa = 9.47KK669 pKa = 9.69VLAVPNALKK678 pKa = 10.98LNIKK682 pKa = 10.46LGVKK686 pKa = 10.14DD687 pKa = 4.02LAPCVFDD694 pKa = 3.37AWYY697 pKa = 10.7NSWLDD702 pKa = 3.99LIWYY706 pKa = 8.15FDD708 pKa = 3.46NYY710 pKa = 10.99LVVSTMKK717 pKa = 10.64DD718 pKa = 3.89YY719 pKa = 10.91ISHH722 pKa = 7.08RR723 pKa = 11.84YY724 pKa = 8.93LRR726 pKa = 11.84RR727 pKa = 11.84QTCYY731 pKa = 10.49QEE733 pKa = 4.45GAMLAYY739 pKa = 9.02RR740 pKa = 11.84TIFSSKK746 pKa = 9.72EE747 pKa = 3.6KK748 pKa = 10.32CLKK751 pKa = 10.45SLFGITSKK759 pKa = 10.84DD760 pKa = 3.58LEE762 pKa = 4.82AMLPMSKK769 pKa = 10.11NGKK772 pKa = 8.3GSNRR776 pKa = 11.84AEE778 pKa = 3.64CSKK781 pKa = 11.12NNKK784 pKa = 7.42TYY786 pKa = 10.6RR787 pKa = 11.84PPKK790 pKa = 9.0STRR793 pKa = 11.84CRR795 pKa = 11.84NSANN799 pKa = 3.3
MM1 pKa = 7.48NPLNQLLSHH10 pKa = 6.66GCSAISSALHH20 pKa = 6.39RR21 pKa = 11.84DD22 pKa = 3.17VAYY25 pKa = 8.84TQISDD30 pKa = 3.66EE31 pKa = 4.62CFDD34 pKa = 3.55SHH36 pKa = 6.35FTQISTEE43 pKa = 4.02YY44 pKa = 10.96SLTTSVRR51 pKa = 11.84KK52 pKa = 9.92LFEE55 pKa = 5.94AYY57 pKa = 8.83MVTMFLKK64 pKa = 10.5KK65 pKa = 9.59HH66 pKa = 5.03MSYY69 pKa = 10.78VFEE72 pKa = 4.88DD73 pKa = 4.92FIGSEE78 pKa = 4.05MGPYY82 pKa = 9.52VQTRR86 pKa = 11.84WRR88 pKa = 11.84IVFDD92 pKa = 3.91LDD94 pKa = 3.65EE95 pKa = 4.6VEE97 pKa = 4.48EE98 pKa = 4.66EE99 pKa = 4.42YY100 pKa = 11.07VYY102 pKa = 11.13KK103 pKa = 10.34PDD105 pKa = 3.98FYY107 pKa = 11.13HH108 pKa = 6.87YY109 pKa = 10.78DD110 pKa = 3.39VYY112 pKa = 10.99DD113 pKa = 3.64YY114 pKa = 11.71EE115 pKa = 4.46MGVDD119 pKa = 3.91IPASIMDD126 pKa = 6.05DD127 pKa = 3.3IPFCEE132 pKa = 4.47DD133 pKa = 3.44AEE135 pKa = 4.35AVTCRR140 pKa = 11.84NCGSFDD146 pKa = 3.52EE147 pKa = 4.86MEE149 pKa = 4.39TVYY152 pKa = 10.58QAATSSDD159 pKa = 3.16EE160 pKa = 4.13SRR162 pKa = 11.84ANVCDD167 pKa = 3.09VDD169 pKa = 4.8APHH172 pKa = 7.23DD173 pKa = 4.1TIKK176 pKa = 10.64WSDD179 pKa = 3.83EE180 pKa = 3.67PDD182 pKa = 3.45LTVDD186 pKa = 5.1DD187 pKa = 4.77PSKK190 pKa = 11.08LHH192 pKa = 7.11DD193 pKa = 5.38LDD195 pKa = 6.56DD196 pKa = 4.87EE197 pKa = 4.87VPQLQLEE204 pKa = 4.57TVGRR208 pKa = 11.84VVQSVEE214 pKa = 3.98PLVPDD219 pKa = 3.45INSFEE224 pKa = 4.32TNQLHH229 pKa = 5.98LVKK232 pKa = 10.4EE233 pKa = 4.43VVLDD237 pKa = 3.66RR238 pKa = 11.84FEE240 pKa = 6.52SILEE244 pKa = 3.99CHH246 pKa = 6.48EE247 pKa = 4.31KK248 pKa = 10.75VLARR252 pKa = 11.84NDD254 pKa = 3.44WKK256 pKa = 11.04SLAMYY261 pKa = 9.95VPTGEE266 pKa = 4.32IPSVSDD272 pKa = 3.06ILEE275 pKa = 4.18IPVKK279 pKa = 9.4HH280 pKa = 5.49TQPEE284 pKa = 4.95FIQMAIDD291 pKa = 3.56EE292 pKa = 4.74MLPGVSDD299 pKa = 3.64FDD301 pKa = 4.42DD302 pKa = 4.65RR303 pKa = 11.84FFQEE307 pKa = 3.89IVEE310 pKa = 4.3TSDD313 pKa = 2.94INLEE317 pKa = 3.73IDD319 pKa = 3.42RR320 pKa = 11.84ASIDD324 pKa = 3.21MSVFNDD330 pKa = 3.32WTPKK334 pKa = 9.87SGRR337 pKa = 11.84LNGLFQTGNISKK349 pKa = 10.21RR350 pKa = 11.84VPTFRR355 pKa = 11.84EE356 pKa = 3.29ASLAIKK362 pKa = 9.92KK363 pKa = 10.38RR364 pKa = 11.84NLNVPDD370 pKa = 4.15LQQVLHH376 pKa = 7.06EE377 pKa = 4.86DD378 pKa = 3.42DD379 pKa = 3.73EE380 pKa = 5.08ARR382 pKa = 11.84FIANKK387 pKa = 10.04FINTVIDD394 pKa = 4.11PNKK397 pKa = 9.5LAQFPGYY404 pKa = 9.77ISEE407 pKa = 4.62GEE409 pKa = 3.54MGYY412 pKa = 10.49FNKK415 pKa = 10.35YY416 pKa = 10.32LSGKK420 pKa = 6.74TVPDD424 pKa = 3.82DD425 pKa = 4.67AFVDD429 pKa = 4.29PCALVSMDD437 pKa = 4.87KK438 pKa = 9.87YY439 pKa = 10.69RR440 pKa = 11.84HH441 pKa = 5.51MIKK444 pKa = 8.45TQLKK448 pKa = 8.28PVEE451 pKa = 4.32DD452 pKa = 3.76TSQLFEE458 pKa = 4.66RR459 pKa = 11.84PLAATITYY467 pKa = 9.23HH468 pKa = 7.77DD469 pKa = 4.3KK470 pKa = 11.19GKK472 pKa = 10.97VMSTSPIFLMMCNRR486 pKa = 11.84LLLCLNDD493 pKa = 4.74KK494 pKa = 10.8ISIPSGKK501 pKa = 8.93HH502 pKa = 4.4HH503 pKa = 6.45QLFSLDD509 pKa = 3.38PFAFEE514 pKa = 3.71MTKK517 pKa = 10.08EE518 pKa = 4.05FKK520 pKa = 10.65EE521 pKa = 3.75IDD523 pKa = 3.38FSKK526 pKa = 10.51FDD528 pKa = 3.39KK529 pKa = 10.68SQQRR533 pKa = 11.84LHH535 pKa = 7.41HH536 pKa = 7.01LIQFHH541 pKa = 6.82IFTALGAPKK550 pKa = 10.28DD551 pKa = 3.96FLDD554 pKa = 3.35MWFASHH560 pKa = 7.23EE561 pKa = 4.05ISHH564 pKa = 6.59IRR566 pKa = 11.84DD567 pKa = 3.92GPCGIGFSVNYY578 pKa = 8.16QRR580 pKa = 11.84RR581 pKa = 11.84TGDD584 pKa = 2.69ACTYY588 pKa = 10.06LGNTIITLSALAYY601 pKa = 9.8MYY603 pKa = 10.97DD604 pKa = 3.7LLDD607 pKa = 4.26PNVTFVIASGDD618 pKa = 3.52DD619 pKa = 3.56SLIGSIKK626 pKa = 10.23PLDD629 pKa = 3.61RR630 pKa = 11.84SEE632 pKa = 4.05EE633 pKa = 4.18FKK635 pKa = 11.03FVTLFNFEE643 pKa = 4.35AKK645 pKa = 10.1FPHH648 pKa = 5.94NQPFVCSKK656 pKa = 9.39FLCLLPTVSGGKK668 pKa = 9.47KK669 pKa = 9.69VLAVPNALKK678 pKa = 10.98LNIKK682 pKa = 10.46LGVKK686 pKa = 10.14DD687 pKa = 4.02LAPCVFDD694 pKa = 3.37AWYY697 pKa = 10.7NSWLDD702 pKa = 3.99LIWYY706 pKa = 8.15FDD708 pKa = 3.46NYY710 pKa = 10.99LVVSTMKK717 pKa = 10.64DD718 pKa = 3.89YY719 pKa = 10.91ISHH722 pKa = 7.08RR723 pKa = 11.84YY724 pKa = 8.93LRR726 pKa = 11.84RR727 pKa = 11.84QTCYY731 pKa = 10.49QEE733 pKa = 4.45GAMLAYY739 pKa = 9.02RR740 pKa = 11.84TIFSSKK746 pKa = 9.72EE747 pKa = 3.6KK748 pKa = 10.32CLKK751 pKa = 10.45SLFGITSKK759 pKa = 10.84DD760 pKa = 3.58LEE762 pKa = 4.82AMLPMSKK769 pKa = 10.11NGKK772 pKa = 8.3GSNRR776 pKa = 11.84AEE778 pKa = 3.64CSKK781 pKa = 11.12NNKK784 pKa = 7.42TYY786 pKa = 10.6RR787 pKa = 11.84PPKK790 pKa = 9.0STRR793 pKa = 11.84CRR795 pKa = 11.84NSANN799 pKa = 3.3
Molecular weight: 91.31 kDa
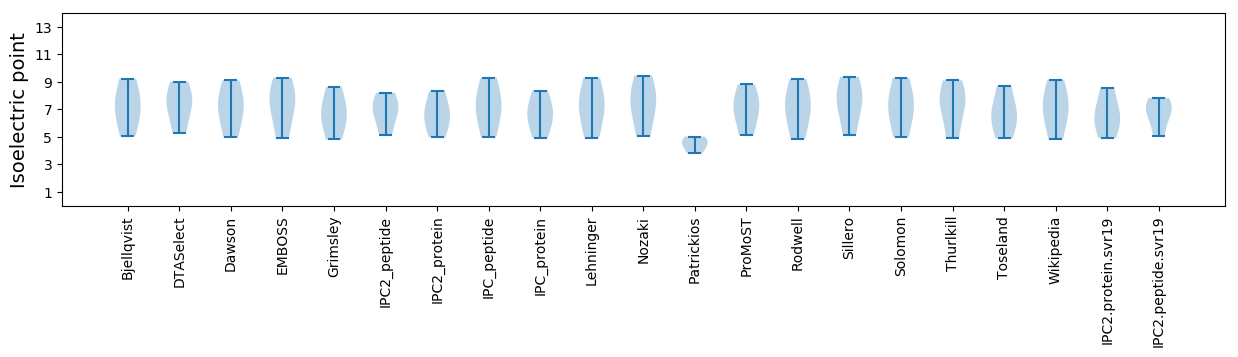
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q98602|Q98602_9BROM Coat protein OS=Prunus necrotic ringspot virus OX=37733 PE=3 SV=1
MM1 pKa = 7.17VCRR4 pKa = 11.84ICNHH8 pKa = 4.64THH10 pKa = 6.79AGGCRR15 pKa = 11.84SCKK18 pKa = 9.53RR19 pKa = 11.84CHH21 pKa = 6.49PNDD24 pKa = 3.44ALVPLRR30 pKa = 11.84AQQRR34 pKa = 11.84AANNPNRR41 pKa = 11.84NRR43 pKa = 11.84NPNRR47 pKa = 11.84VSSGIGPAVRR57 pKa = 11.84PQPVVKK63 pKa = 8.13TTWTVRR69 pKa = 11.84GPNVPPRR76 pKa = 11.84IPKK79 pKa = 10.17GYY81 pKa = 8.25VAHH84 pKa = 6.08NHH86 pKa = 6.2RR87 pKa = 11.84EE88 pKa = 4.16VTTTEE93 pKa = 3.58AVKK96 pKa = 10.79YY97 pKa = 10.23LSIDD101 pKa = 3.41FTTTLPQLMGQNLTLLTVIVRR122 pKa = 11.84MNSMSSNGWIGMVEE136 pKa = 4.39DD137 pKa = 4.33YY138 pKa = 11.01KK139 pKa = 11.23VDD141 pKa = 3.88QPDD144 pKa = 3.77GPNALSRR151 pKa = 11.84KK152 pKa = 9.54GFLKK156 pKa = 10.31DD157 pKa = 4.37QPRR160 pKa = 11.84GWQFEE165 pKa = 4.36PPSDD169 pKa = 4.31LDD171 pKa = 3.35FDD173 pKa = 4.23TFARR177 pKa = 11.84THH179 pKa = 5.58RR180 pKa = 11.84VVIEE184 pKa = 4.53FKK186 pKa = 10.65TEE188 pKa = 3.59VPAGAKK194 pKa = 10.02VLVRR198 pKa = 11.84DD199 pKa = 4.43LYY201 pKa = 11.58VVVSDD206 pKa = 4.51LPRR209 pKa = 11.84VQIPTDD215 pKa = 3.44VLLVDD220 pKa = 4.27EE221 pKa = 5.62DD222 pKa = 4.01LLEE225 pKa = 4.15II226 pKa = 5.03
MM1 pKa = 7.17VCRR4 pKa = 11.84ICNHH8 pKa = 4.64THH10 pKa = 6.79AGGCRR15 pKa = 11.84SCKK18 pKa = 9.53RR19 pKa = 11.84CHH21 pKa = 6.49PNDD24 pKa = 3.44ALVPLRR30 pKa = 11.84AQQRR34 pKa = 11.84AANNPNRR41 pKa = 11.84NRR43 pKa = 11.84NPNRR47 pKa = 11.84VSSGIGPAVRR57 pKa = 11.84PQPVVKK63 pKa = 8.13TTWTVRR69 pKa = 11.84GPNVPPRR76 pKa = 11.84IPKK79 pKa = 10.17GYY81 pKa = 8.25VAHH84 pKa = 6.08NHH86 pKa = 6.2RR87 pKa = 11.84EE88 pKa = 4.16VTTTEE93 pKa = 3.58AVKK96 pKa = 10.79YY97 pKa = 10.23LSIDD101 pKa = 3.41FTTTLPQLMGQNLTLLTVIVRR122 pKa = 11.84MNSMSSNGWIGMVEE136 pKa = 4.39DD137 pKa = 4.33YY138 pKa = 11.01KK139 pKa = 11.23VDD141 pKa = 3.88QPDD144 pKa = 3.77GPNALSRR151 pKa = 11.84KK152 pKa = 9.54GFLKK156 pKa = 10.31DD157 pKa = 4.37QPRR160 pKa = 11.84GWQFEE165 pKa = 4.36PPSDD169 pKa = 4.31LDD171 pKa = 3.35FDD173 pKa = 4.23TFARR177 pKa = 11.84THH179 pKa = 5.58RR180 pKa = 11.84VVIEE184 pKa = 4.53FKK186 pKa = 10.65TEE188 pKa = 3.59VPAGAKK194 pKa = 10.02VLVRR198 pKa = 11.84DD199 pKa = 4.43LYY201 pKa = 11.58VVVSDD206 pKa = 4.51LPRR209 pKa = 11.84VQIPTDD215 pKa = 3.44VLLVDD220 pKa = 4.27EE221 pKa = 5.62DD222 pKa = 4.01LLEE225 pKa = 4.15II226 pKa = 5.03
Molecular weight: 25.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2351 |
226 |
1045 |
587.8 |
66.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.168 ± 0.675 | 2.169 ± 0.105 |
7.188 ± 0.503 | 6.04 ± 0.381 |
4.296 ± 0.87 | 4.977 ± 0.443 |
2.382 ± 0.21 | 5.359 ± 0.382 |
6.125 ± 0.357 | 8.677 ± 0.157 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.807 ± 0.339 | 3.828 ± 0.589 |
4.934 ± 0.679 | 3.105 ± 0.172 |
5.955 ± 0.924 | 7.741 ± 0.496 |
5.955 ± 0.495 | 8.124 ± 0.832 |
1.063 ± 0.073 | 3.105 ± 0.422 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
