
Prevotella sp. CAG:1124
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Prevotellaceae; Prevotella; environmental samples
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
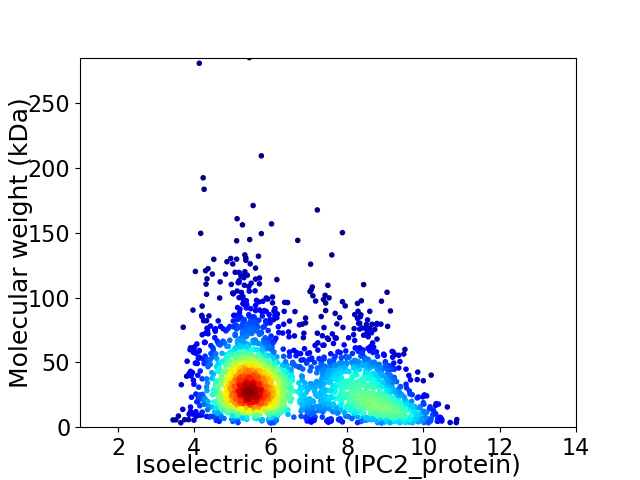
Virtual 2D-PAGE plot for 2441 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5KZL9|R5KZL9_9BACT GHMP kinase protein OS=Prevotella sp. CAG:1124 OX=1262920 GN=BN467_00759 PE=4 SV=1
MM1 pKa = 7.58KK2 pKa = 10.51KK3 pKa = 10.54LFLTAMAFVAMMSAMSFTSCSDD25 pKa = 4.06DD26 pKa = 3.45DD27 pKa = 4.28TNDD30 pKa = 3.28NGGGGGGGSTTGGVEE45 pKa = 3.92LNGAIEE51 pKa = 4.32GTVTLEE57 pKa = 3.42ADD59 pKa = 3.06KK60 pKa = 10.68EE61 pKa = 4.17YY62 pKa = 10.97RR63 pKa = 11.84LTGAVTVPDD72 pKa = 4.31GATLNIPAGTTIRR85 pKa = 11.84ATQGFSSYY93 pKa = 11.04ILVEE97 pKa = 4.05RR98 pKa = 11.84GGTINAEE105 pKa = 4.03GTADD109 pKa = 3.52KK110 pKa = 10.57PIVFTADD117 pKa = 3.01TDD119 pKa = 3.82NATAGYY125 pKa = 8.74WGGLIINGNAPITGGGEE142 pKa = 4.19GVTEE146 pKa = 3.72IDD148 pKa = 3.56NNVSYY153 pKa = 11.02GGNDD157 pKa = 3.32ANDD160 pKa = 3.25NSGVIRR166 pKa = 11.84YY167 pKa = 9.18VEE169 pKa = 4.08LWYY172 pKa = 9.77TGARR176 pKa = 11.84SSADD180 pKa = 3.05IEE182 pKa = 4.94HH183 pKa = 7.07NGLTLNGVGNGTTIEE198 pKa = 4.05NVYY201 pKa = 9.94IAEE204 pKa = 4.43GADD207 pKa = 3.41DD208 pKa = 4.88GIEE211 pKa = 4.19FFGGTVNVSNLLVVNCDD228 pKa = 3.5DD229 pKa = 4.56DD230 pKa = 6.25CFDD233 pKa = 3.77FTQGYY238 pKa = 10.06SGTLTNCYY246 pKa = 10.0GIWEE250 pKa = 4.92DD251 pKa = 4.19GFTSTEE257 pKa = 3.67EE258 pKa = 3.97DD259 pKa = 3.46PRR261 pKa = 11.84GVEE264 pKa = 4.42ADD266 pKa = 3.64GNLDD270 pKa = 3.05GDD272 pKa = 4.4MPDD275 pKa = 4.42GAPQADD281 pKa = 3.85FTINGMTIVNNSSTQSMQDD300 pKa = 3.15AIKK303 pKa = 10.35VRR305 pKa = 11.84RR306 pKa = 11.84GATAHH311 pKa = 6.01ITDD314 pKa = 3.54AVVYY318 pKa = 10.33GSGQIEE324 pKa = 4.23NLIDD328 pKa = 3.56LTDD331 pKa = 3.78GKK333 pKa = 10.91GGAGRR338 pKa = 11.84STDD341 pKa = 3.09MSVSNNAYY349 pKa = 9.59YY350 pKa = 10.18ISDD353 pKa = 3.33ATINDD358 pKa = 3.49GGTTYY363 pKa = 10.88SGVNVEE369 pKa = 4.56TGNDD373 pKa = 4.0GADD376 pKa = 3.23VSALSWTGYY385 pKa = 9.29EE386 pKa = 4.96FPQLNNTTVAAAGDD400 pKa = 4.07LPLEE404 pKa = 4.3ISSPIALEE412 pKa = 4.04AGQEE416 pKa = 4.06YY417 pKa = 9.91FINGSVHH424 pKa = 5.15VKK426 pKa = 10.24EE427 pKa = 4.91GGVLLIPAGMTIKK440 pKa = 10.54ARR442 pKa = 11.84EE443 pKa = 4.13GFSNFILVEE452 pKa = 4.15RR453 pKa = 11.84GGKK456 pKa = 9.36IYY458 pKa = 9.88ATGRR462 pKa = 11.84EE463 pKa = 4.11DD464 pKa = 4.5APITFTADD472 pKa = 2.8ASNATAGYY480 pKa = 8.41WGGIIINGNAVISGPSGSVNEE501 pKa = 4.63GVTEE505 pKa = 3.76IDD507 pKa = 3.23NNVPYY512 pKa = 10.9GGDD515 pKa = 3.39QNNDD519 pKa = 2.72NSGVLTYY526 pKa = 10.55VSILYY531 pKa = 10.48SGARR535 pKa = 11.84SSADD539 pKa = 3.0IEE541 pKa = 4.94HH542 pKa = 7.07NGLTLNGVGAGTTIEE557 pKa = 4.38NIYY560 pKa = 9.78IAEE563 pKa = 4.37GADD566 pKa = 3.45DD567 pKa = 4.88GIEE570 pKa = 4.08FFGGSVNVSNLLVVNCDD587 pKa = 3.5DD588 pKa = 4.56DD589 pKa = 6.25CFDD592 pKa = 3.77FTQGYY597 pKa = 10.06SGTLTNCYY605 pKa = 9.78GRR607 pKa = 11.84WEE609 pKa = 4.34AGFTSTEE616 pKa = 3.72LDD618 pKa = 3.47PRR620 pKa = 11.84GVEE623 pKa = 4.22ADD625 pKa = 3.62GNLDD629 pKa = 3.79GEE631 pKa = 4.97GSTAHH636 pKa = 6.59SPQSDD641 pKa = 3.55FTIEE645 pKa = 3.99NMTIEE650 pKa = 4.47NLSSYY655 pKa = 10.44PMQDD659 pKa = 2.97AIKK662 pKa = 10.34VRR664 pKa = 11.84RR665 pKa = 11.84LAKK668 pKa = 9.66ATINNALVIGSGEE681 pKa = 3.86IQEE684 pKa = 5.51LVDD687 pKa = 4.19LTDD690 pKa = 4.42GLDD693 pKa = 3.92DD694 pKa = 5.39ADD696 pKa = 3.91PTSVINVTNGATNVGAQTNSSNPEE720 pKa = 3.65GATVNIADD728 pKa = 4.43GNTGCEE734 pKa = 3.58TDD736 pKa = 2.87IFGWTGYY743 pKa = 7.51TLL745 pKa = 3.63
MM1 pKa = 7.58KK2 pKa = 10.51KK3 pKa = 10.54LFLTAMAFVAMMSAMSFTSCSDD25 pKa = 4.06DD26 pKa = 3.45DD27 pKa = 4.28TNDD30 pKa = 3.28NGGGGGGGSTTGGVEE45 pKa = 3.92LNGAIEE51 pKa = 4.32GTVTLEE57 pKa = 3.42ADD59 pKa = 3.06KK60 pKa = 10.68EE61 pKa = 4.17YY62 pKa = 10.97RR63 pKa = 11.84LTGAVTVPDD72 pKa = 4.31GATLNIPAGTTIRR85 pKa = 11.84ATQGFSSYY93 pKa = 11.04ILVEE97 pKa = 4.05RR98 pKa = 11.84GGTINAEE105 pKa = 4.03GTADD109 pKa = 3.52KK110 pKa = 10.57PIVFTADD117 pKa = 3.01TDD119 pKa = 3.82NATAGYY125 pKa = 8.74WGGLIINGNAPITGGGEE142 pKa = 4.19GVTEE146 pKa = 3.72IDD148 pKa = 3.56NNVSYY153 pKa = 11.02GGNDD157 pKa = 3.32ANDD160 pKa = 3.25NSGVIRR166 pKa = 11.84YY167 pKa = 9.18VEE169 pKa = 4.08LWYY172 pKa = 9.77TGARR176 pKa = 11.84SSADD180 pKa = 3.05IEE182 pKa = 4.94HH183 pKa = 7.07NGLTLNGVGNGTTIEE198 pKa = 4.05NVYY201 pKa = 9.94IAEE204 pKa = 4.43GADD207 pKa = 3.41DD208 pKa = 4.88GIEE211 pKa = 4.19FFGGTVNVSNLLVVNCDD228 pKa = 3.5DD229 pKa = 4.56DD230 pKa = 6.25CFDD233 pKa = 3.77FTQGYY238 pKa = 10.06SGTLTNCYY246 pKa = 10.0GIWEE250 pKa = 4.92DD251 pKa = 4.19GFTSTEE257 pKa = 3.67EE258 pKa = 3.97DD259 pKa = 3.46PRR261 pKa = 11.84GVEE264 pKa = 4.42ADD266 pKa = 3.64GNLDD270 pKa = 3.05GDD272 pKa = 4.4MPDD275 pKa = 4.42GAPQADD281 pKa = 3.85FTINGMTIVNNSSTQSMQDD300 pKa = 3.15AIKK303 pKa = 10.35VRR305 pKa = 11.84RR306 pKa = 11.84GATAHH311 pKa = 6.01ITDD314 pKa = 3.54AVVYY318 pKa = 10.33GSGQIEE324 pKa = 4.23NLIDD328 pKa = 3.56LTDD331 pKa = 3.78GKK333 pKa = 10.91GGAGRR338 pKa = 11.84STDD341 pKa = 3.09MSVSNNAYY349 pKa = 9.59YY350 pKa = 10.18ISDD353 pKa = 3.33ATINDD358 pKa = 3.49GGTTYY363 pKa = 10.88SGVNVEE369 pKa = 4.56TGNDD373 pKa = 4.0GADD376 pKa = 3.23VSALSWTGYY385 pKa = 9.29EE386 pKa = 4.96FPQLNNTTVAAAGDD400 pKa = 4.07LPLEE404 pKa = 4.3ISSPIALEE412 pKa = 4.04AGQEE416 pKa = 4.06YY417 pKa = 9.91FINGSVHH424 pKa = 5.15VKK426 pKa = 10.24EE427 pKa = 4.91GGVLLIPAGMTIKK440 pKa = 10.54ARR442 pKa = 11.84EE443 pKa = 4.13GFSNFILVEE452 pKa = 4.15RR453 pKa = 11.84GGKK456 pKa = 9.36IYY458 pKa = 9.88ATGRR462 pKa = 11.84EE463 pKa = 4.11DD464 pKa = 4.5APITFTADD472 pKa = 2.8ASNATAGYY480 pKa = 8.41WGGIIINGNAVISGPSGSVNEE501 pKa = 4.63GVTEE505 pKa = 3.76IDD507 pKa = 3.23NNVPYY512 pKa = 10.9GGDD515 pKa = 3.39QNNDD519 pKa = 2.72NSGVLTYY526 pKa = 10.55VSILYY531 pKa = 10.48SGARR535 pKa = 11.84SSADD539 pKa = 3.0IEE541 pKa = 4.94HH542 pKa = 7.07NGLTLNGVGAGTTIEE557 pKa = 4.38NIYY560 pKa = 9.78IAEE563 pKa = 4.37GADD566 pKa = 3.45DD567 pKa = 4.88GIEE570 pKa = 4.08FFGGSVNVSNLLVVNCDD587 pKa = 3.5DD588 pKa = 4.56DD589 pKa = 6.25CFDD592 pKa = 3.77FTQGYY597 pKa = 10.06SGTLTNCYY605 pKa = 9.78GRR607 pKa = 11.84WEE609 pKa = 4.34AGFTSTEE616 pKa = 3.72LDD618 pKa = 3.47PRR620 pKa = 11.84GVEE623 pKa = 4.22ADD625 pKa = 3.62GNLDD629 pKa = 3.79GEE631 pKa = 4.97GSTAHH636 pKa = 6.59SPQSDD641 pKa = 3.55FTIEE645 pKa = 3.99NMTIEE650 pKa = 4.47NLSSYY655 pKa = 10.44PMQDD659 pKa = 2.97AIKK662 pKa = 10.34VRR664 pKa = 11.84RR665 pKa = 11.84LAKK668 pKa = 9.66ATINNALVIGSGEE681 pKa = 3.86IQEE684 pKa = 5.51LVDD687 pKa = 4.19LTDD690 pKa = 4.42GLDD693 pKa = 3.92DD694 pKa = 5.39ADD696 pKa = 3.91PTSVINVTNGATNVGAQTNSSNPEE720 pKa = 3.65GATVNIADD728 pKa = 4.43GNTGCEE734 pKa = 3.58TDD736 pKa = 2.87IFGWTGYY743 pKa = 7.51TLL745 pKa = 3.63
Molecular weight: 77.18 kDa
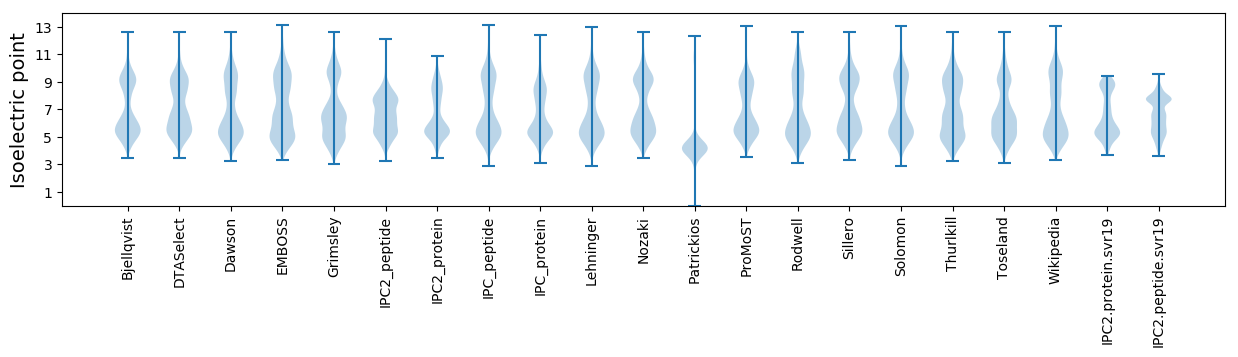
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5KK40|R5KK40_9BACT Uncharacterized protein OS=Prevotella sp. CAG:1124 OX=1262920 GN=BN467_02137 PE=4 SV=1
MM1 pKa = 7.73PMLIVLATTTLGRR14 pKa = 11.84WRR16 pKa = 11.84SHH18 pKa = 5.57RR19 pKa = 11.84RR20 pKa = 11.84PLTRR24 pKa = 11.84LPATMPKK31 pKa = 9.24PAAIIKK37 pKa = 10.28AGSTSAAKK45 pKa = 10.01PPSVVSKK52 pKa = 11.33GLMM55 pKa = 3.3
MM1 pKa = 7.73PMLIVLATTTLGRR14 pKa = 11.84WRR16 pKa = 11.84SHH18 pKa = 5.57RR19 pKa = 11.84RR20 pKa = 11.84PLTRR24 pKa = 11.84LPATMPKK31 pKa = 9.24PAAIIKK37 pKa = 10.28AGSTSAAKK45 pKa = 10.01PPSVVSKK52 pKa = 11.33GLMM55 pKa = 3.3
Molecular weight: 5.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
807868 |
29 |
2609 |
331.0 |
37.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.646 ± 0.046 | 1.408 ± 0.022 |
6.19 ± 0.044 | 5.966 ± 0.046 |
4.29 ± 0.031 | 7.313 ± 0.044 |
1.881 ± 0.021 | 6.541 ± 0.045 |
6.069 ± 0.05 | 8.545 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.016 ± 0.028 | 4.989 ± 0.043 |
3.708 ± 0.027 | 2.992 ± 0.028 |
5.15 ± 0.048 | 5.91 ± 0.052 |
5.886 ± 0.045 | 6.954 ± 0.045 |
1.212 ± 0.019 | 4.332 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
