
Phytophthora rubi
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Oomycota; Peronosporales; Peronosporaceae; Phytophthora
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
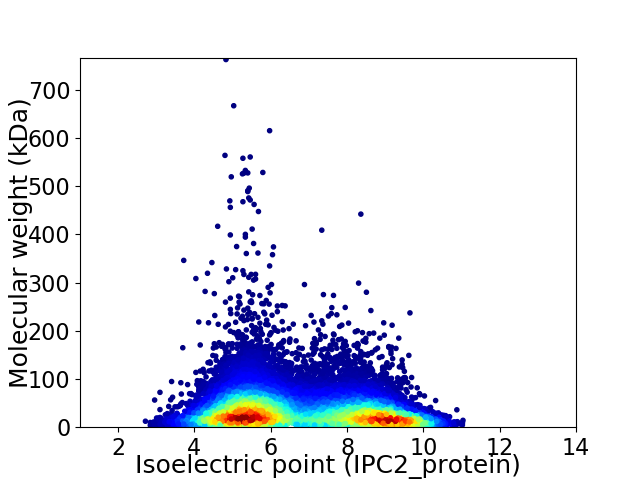
Virtual 2D-PAGE plot for 35379 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6A4C5U4|A0A6A4C5U4_9STRA Uncharacterized protein OS=Phytophthora rubi OX=129364 GN=PR002_g26572 PE=4 SV=1
MM1 pKa = 7.44TFLILEE7 pKa = 4.28NSSYY11 pKa = 9.77STSSQQPVYY20 pKa = 10.81LCVDD24 pKa = 3.74MNIHH28 pKa = 6.83ALLLVAASMTASATAKK44 pKa = 9.17FTVTSGTTEE53 pKa = 3.83ATTSTTEE60 pKa = 3.39NVAFVKK66 pKa = 10.31QWTLNSATTADD77 pKa = 4.32TIDD80 pKa = 5.43SIDD83 pKa = 4.08LDD85 pKa = 4.04LAGRR89 pKa = 11.84VYY91 pKa = 10.86VSYY94 pKa = 11.41VSGLPSGVLGYY105 pKa = 11.23VNVSGDD111 pKa = 3.4SQTVVDD117 pKa = 5.39AVTVINDD124 pKa = 3.96DD125 pKa = 5.31ANDD128 pKa = 3.77TDD130 pKa = 6.41DD131 pKa = 6.07NDD133 pKa = 4.24TNDD136 pKa = 3.84NDD138 pKa = 3.95GEE140 pKa = 4.59LNVRR144 pKa = 11.84IGNSTSTATTLSGYY158 pKa = 10.66LLTEE162 pKa = 5.02IILASSGVVTDD173 pKa = 3.82VKK175 pKa = 10.3SQRR178 pKa = 11.84SAQVVVEE185 pKa = 4.38DD186 pKa = 4.42GVLVSSSTTAEE197 pKa = 4.15LQVEE201 pKa = 4.55ASGSSAVYY209 pKa = 10.06VSAASTAVSVRR220 pKa = 11.84QLQLDD225 pKa = 3.82AAGTAQLQFNVKK237 pKa = 9.39SVSVTDD243 pKa = 3.57EE244 pKa = 4.16AQFEE248 pKa = 4.51AQGSAVVSVLASSLEE263 pKa = 3.79ASQLDD268 pKa = 4.89LEE270 pKa = 4.65TQSTGTICISAQQVTATNYY289 pKa = 9.59QGQDD293 pKa = 2.86ASRR296 pKa = 11.84ISMPNASSKK305 pKa = 10.83YY306 pKa = 10.21DD307 pKa = 3.49STGSLACDD315 pKa = 3.39EE316 pKa = 4.63SSIPTRR322 pKa = 11.84EE323 pKa = 4.0PSCVSSACASSSTSGTSTTATVVTSAPTTTNDD355 pKa = 4.33DD356 pKa = 4.02DD357 pKa = 6.48ASDD360 pKa = 4.49DD361 pKa = 4.26SNASSTPRR369 pKa = 11.84LAALVVAVIGVVTAGLLL386 pKa = 3.53
MM1 pKa = 7.44TFLILEE7 pKa = 4.28NSSYY11 pKa = 9.77STSSQQPVYY20 pKa = 10.81LCVDD24 pKa = 3.74MNIHH28 pKa = 6.83ALLLVAASMTASATAKK44 pKa = 9.17FTVTSGTTEE53 pKa = 3.83ATTSTTEE60 pKa = 3.39NVAFVKK66 pKa = 10.31QWTLNSATTADD77 pKa = 4.32TIDD80 pKa = 5.43SIDD83 pKa = 4.08LDD85 pKa = 4.04LAGRR89 pKa = 11.84VYY91 pKa = 10.86VSYY94 pKa = 11.41VSGLPSGVLGYY105 pKa = 11.23VNVSGDD111 pKa = 3.4SQTVVDD117 pKa = 5.39AVTVINDD124 pKa = 3.96DD125 pKa = 5.31ANDD128 pKa = 3.77TDD130 pKa = 6.41DD131 pKa = 6.07NDD133 pKa = 4.24TNDD136 pKa = 3.84NDD138 pKa = 3.95GEE140 pKa = 4.59LNVRR144 pKa = 11.84IGNSTSTATTLSGYY158 pKa = 10.66LLTEE162 pKa = 5.02IILASSGVVTDD173 pKa = 3.82VKK175 pKa = 10.3SQRR178 pKa = 11.84SAQVVVEE185 pKa = 4.38DD186 pKa = 4.42GVLVSSSTTAEE197 pKa = 4.15LQVEE201 pKa = 4.55ASGSSAVYY209 pKa = 10.06VSAASTAVSVRR220 pKa = 11.84QLQLDD225 pKa = 3.82AAGTAQLQFNVKK237 pKa = 9.39SVSVTDD243 pKa = 3.57EE244 pKa = 4.16AQFEE248 pKa = 4.51AQGSAVVSVLASSLEE263 pKa = 3.79ASQLDD268 pKa = 4.89LEE270 pKa = 4.65TQSTGTICISAQQVTATNYY289 pKa = 9.59QGQDD293 pKa = 2.86ASRR296 pKa = 11.84ISMPNASSKK305 pKa = 10.83YY306 pKa = 10.21DD307 pKa = 3.49STGSLACDD315 pKa = 3.39EE316 pKa = 4.63SSIPTRR322 pKa = 11.84EE323 pKa = 4.0PSCVSSACASSSTSGTSTTATVVTSAPTTTNDD355 pKa = 4.33DD356 pKa = 4.02DD357 pKa = 6.48ASDD360 pKa = 4.49DD361 pKa = 4.26SNASSTPRR369 pKa = 11.84LAALVVAVIGVVTAGLLL386 pKa = 3.53
Molecular weight: 39.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6A4BL47|A0A6A4BL47_9STRA Chromo domain-containing protein OS=Phytophthora rubi OX=129364 GN=PR003_g29016 PE=4 SV=1
MM1 pKa = 7.77SSQKK5 pKa = 10.26KK6 pKa = 10.05SKK8 pKa = 10.72SQMKK12 pKa = 9.72PKK14 pKa = 10.12SQRR17 pKa = 11.84LSSRR21 pKa = 11.84KK22 pKa = 8.32KK23 pKa = 8.07SKK25 pKa = 9.78RR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 7.38VSKK31 pKa = 9.92RR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 8.37SKK36 pKa = 10.78RR37 pKa = 11.84KK38 pKa = 8.0ISKK41 pKa = 9.53SQVVTT46 pKa = 3.85
MM1 pKa = 7.77SSQKK5 pKa = 10.26KK6 pKa = 10.05SKK8 pKa = 10.72SQMKK12 pKa = 9.72PKK14 pKa = 10.12SQRR17 pKa = 11.84LSSRR21 pKa = 11.84KK22 pKa = 8.32KK23 pKa = 8.07SKK25 pKa = 9.78RR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 7.38VSKK31 pKa = 9.92RR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 8.37SKK36 pKa = 10.78RR37 pKa = 11.84KK38 pKa = 8.0ISKK41 pKa = 9.53SQVVTT46 pKa = 3.85
Molecular weight: 5.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
12151655 |
10 |
10508 |
343.5 |
38.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.602 ± 0.015 | 1.629 ± 0.008 |
5.819 ± 0.01 | 6.625 ± 0.015 |
3.464 ± 0.009 | 6.205 ± 0.015 |
2.321 ± 0.006 | 3.673 ± 0.01 |
4.909 ± 0.016 | 9.097 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.395 ± 0.006 | 3.212 ± 0.008 |
5.112 ± 0.014 | 4.157 ± 0.01 |
6.791 ± 0.016 | 8.151 ± 0.017 |
5.907 ± 0.014 | 7.181 ± 0.014 |
1.277 ± 0.004 | 2.472 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
