
Avon-Heathcote Estuary associated circular virus 12
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 5.01
Get precalculated fractions of proteins
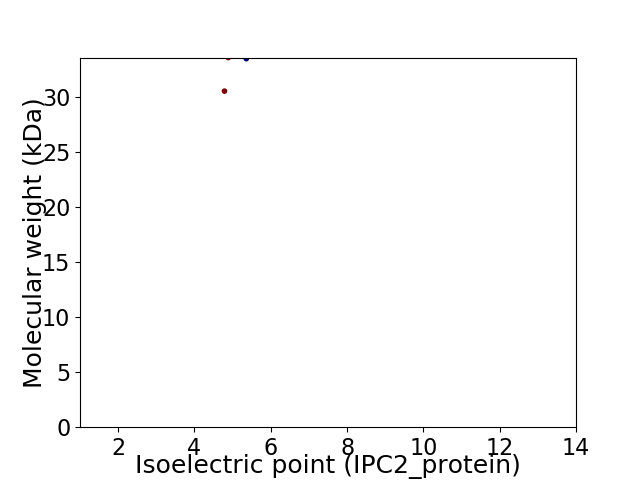
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
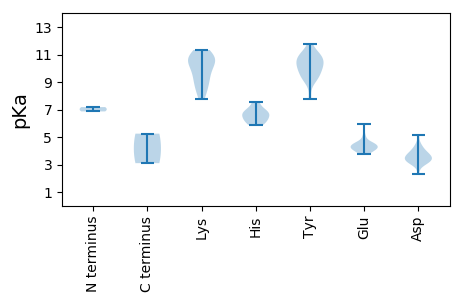
Protein with the lowest isoelectric point:
>tr|A0A0C5IMF1|A0A0C5IMF1_9CIRC Replication-associated protein OS=Avon-Heathcote Estuary associated circular virus 12 OX=1618235 PE=4 SV=1
MM1 pKa = 7.18QPLGLTMSGKK11 pKa = 10.28AKK13 pKa = 8.8QTINEE18 pKa = 3.96FDD20 pKa = 3.13IKK22 pKa = 11.23GNNTLVGVPQMAVMDD37 pKa = 4.14TAYY40 pKa = 10.76CLSKK44 pKa = 10.73LNHH47 pKa = 6.06RR48 pKa = 11.84LYY50 pKa = 10.69RR51 pKa = 11.84QGRR54 pKa = 11.84AYY56 pKa = 9.04TVRR59 pKa = 11.84VEE61 pKa = 4.2ANQDD65 pKa = 3.36SFGSTEE71 pKa = 4.21SIEE74 pKa = 4.62VFALMPTWYY83 pKa = 10.1LRR85 pKa = 11.84SAWRR89 pKa = 11.84MPKK92 pKa = 9.89KK93 pKa = 10.63AYY95 pKa = 10.24DD96 pKa = 3.43EE97 pKa = 4.81AMADD101 pKa = 3.88EE102 pKa = 4.83LASLNPEE109 pKa = 3.82NVARR113 pKa = 11.84WRR115 pKa = 11.84DD116 pKa = 3.52FRR118 pKa = 11.84VGSGITLTGTTDD130 pKa = 3.13ALEE133 pKa = 4.43FGGAVPNLWQLDD145 pKa = 3.99EE146 pKa = 4.47TGVAPVRR153 pKa = 11.84TPFTAGEE160 pKa = 4.22YY161 pKa = 9.39NLSYY165 pKa = 11.09AHH167 pKa = 6.84NLDD170 pKa = 3.68TGVNMQWWLKK180 pKa = 10.97GGDD183 pKa = 3.41ATHH186 pKa = 6.69FGVFEE191 pKa = 4.49EE192 pKa = 4.59YY193 pKa = 10.41SKK195 pKa = 11.33SRR197 pKa = 11.84NEE199 pKa = 4.07SASPEE204 pKa = 4.13TVIQSMPYY212 pKa = 9.33EE213 pKa = 4.09ALEE216 pKa = 4.56AEE218 pKa = 5.11GEE220 pKa = 4.19DD221 pKa = 3.49EE222 pKa = 5.97DD223 pKa = 5.15YY224 pKa = 11.78EE225 pKa = 4.33EE226 pKa = 4.34VQANGNEE233 pKa = 3.99PPYY236 pKa = 10.37NANAFPNGIWVRR248 pKa = 11.84VGVLEE253 pKa = 4.56KK254 pKa = 10.31STSLTQRR261 pKa = 11.84STGFFHH267 pKa = 7.54APLGMVAFF275 pKa = 5.25
MM1 pKa = 7.18QPLGLTMSGKK11 pKa = 10.28AKK13 pKa = 8.8QTINEE18 pKa = 3.96FDD20 pKa = 3.13IKK22 pKa = 11.23GNNTLVGVPQMAVMDD37 pKa = 4.14TAYY40 pKa = 10.76CLSKK44 pKa = 10.73LNHH47 pKa = 6.06RR48 pKa = 11.84LYY50 pKa = 10.69RR51 pKa = 11.84QGRR54 pKa = 11.84AYY56 pKa = 9.04TVRR59 pKa = 11.84VEE61 pKa = 4.2ANQDD65 pKa = 3.36SFGSTEE71 pKa = 4.21SIEE74 pKa = 4.62VFALMPTWYY83 pKa = 10.1LRR85 pKa = 11.84SAWRR89 pKa = 11.84MPKK92 pKa = 9.89KK93 pKa = 10.63AYY95 pKa = 10.24DD96 pKa = 3.43EE97 pKa = 4.81AMADD101 pKa = 3.88EE102 pKa = 4.83LASLNPEE109 pKa = 3.82NVARR113 pKa = 11.84WRR115 pKa = 11.84DD116 pKa = 3.52FRR118 pKa = 11.84VGSGITLTGTTDD130 pKa = 3.13ALEE133 pKa = 4.43FGGAVPNLWQLDD145 pKa = 3.99EE146 pKa = 4.47TGVAPVRR153 pKa = 11.84TPFTAGEE160 pKa = 4.22YY161 pKa = 9.39NLSYY165 pKa = 11.09AHH167 pKa = 6.84NLDD170 pKa = 3.68TGVNMQWWLKK180 pKa = 10.97GGDD183 pKa = 3.41ATHH186 pKa = 6.69FGVFEE191 pKa = 4.49EE192 pKa = 4.59YY193 pKa = 10.41SKK195 pKa = 11.33SRR197 pKa = 11.84NEE199 pKa = 4.07SASPEE204 pKa = 4.13TVIQSMPYY212 pKa = 9.33EE213 pKa = 4.09ALEE216 pKa = 4.56AEE218 pKa = 5.11GEE220 pKa = 4.19DD221 pKa = 3.49EE222 pKa = 5.97DD223 pKa = 5.15YY224 pKa = 11.78EE225 pKa = 4.33EE226 pKa = 4.34VQANGNEE233 pKa = 3.99PPYY236 pKa = 10.37NANAFPNGIWVRR248 pKa = 11.84VGVLEE253 pKa = 4.56KK254 pKa = 10.31STSLTQRR261 pKa = 11.84STGFFHH267 pKa = 7.54APLGMVAFF275 pKa = 5.25
Molecular weight: 30.56 kDa
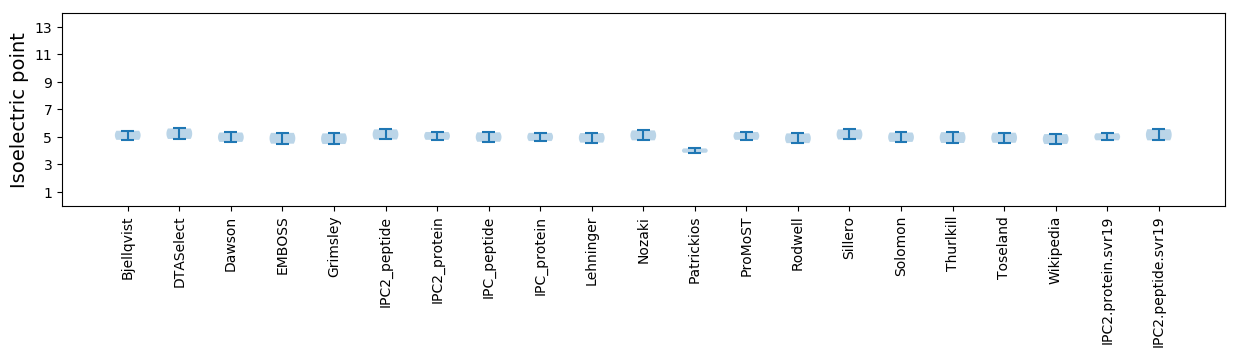
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IMF1|A0A0C5IMF1_9CIRC Replication-associated protein OS=Avon-Heathcote Estuary associated circular virus 12 OX=1618235 PE=4 SV=1
MM1 pKa = 6.86SQRR4 pKa = 11.84TRR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 10.08CLTLNNPTDD17 pKa = 3.69AEE19 pKa = 4.28IASLLDD25 pKa = 3.6VQEE28 pKa = 4.52GTLKK32 pKa = 10.64RR33 pKa = 11.84GFIALEE39 pKa = 4.1VGAEE43 pKa = 4.26GTPHH47 pKa = 6.29LQGFVHH53 pKa = 6.96LKK55 pKa = 8.71NAKK58 pKa = 9.21TGTALKK64 pKa = 10.64KK65 pKa = 10.15MLGSDD70 pKa = 2.95RR71 pKa = 11.84WHH73 pKa = 6.36WGKK76 pKa = 11.0ANGTDD81 pKa = 3.96FEE83 pKa = 4.22NWAYY87 pKa = 10.07IMEE90 pKa = 4.52DD91 pKa = 3.25VEE93 pKa = 5.57GKK95 pKa = 9.9VQGDD99 pKa = 4.48LLIQWGDD106 pKa = 3.24KK107 pKa = 9.61PAEE110 pKa = 4.29EE111 pKa = 5.08GEE113 pKa = 3.94PDD115 pKa = 3.19AWDD118 pKa = 4.45SILMMIEE125 pKa = 3.84AGEE128 pKa = 4.2DD129 pKa = 2.97DD130 pKa = 3.69RR131 pKa = 11.84AIVRR135 pKa = 11.84KK136 pKa = 8.39WPSIAIRR143 pKa = 11.84CQSAIAKK150 pKa = 9.36YY151 pKa = 9.65RR152 pKa = 11.84ASFEE156 pKa = 4.16WAEE159 pKa = 3.89CRR161 pKa = 11.84AWRR164 pKa = 11.84EE165 pKa = 3.79VEE167 pKa = 3.89VEE169 pKa = 4.38YY170 pKa = 10.75ISGPTGCGKK179 pKa = 8.2TRR181 pKa = 11.84EE182 pKa = 4.19ALYY185 pKa = 10.97LPDD188 pKa = 3.69GRR190 pKa = 11.84VNTDD194 pKa = 2.31VYY196 pKa = 11.0RR197 pKa = 11.84CTNGKK202 pKa = 9.16HH203 pKa = 5.88PFDD206 pKa = 4.18MYY208 pKa = 10.91DD209 pKa = 3.42GEE211 pKa = 4.42GTIVFEE217 pKa = 4.61EE218 pKa = 4.53FRR220 pKa = 11.84SQYY223 pKa = 7.73TCRR226 pKa = 11.84DD227 pKa = 3.19MLNWVDD233 pKa = 3.92GHH235 pKa = 6.69PLMLPARR242 pKa = 11.84YY243 pKa = 9.24ADD245 pKa = 3.61RR246 pKa = 11.84MAKK249 pKa = 7.73FTKK252 pKa = 10.51VIILSNWRR260 pKa = 11.84FEE262 pKa = 4.15EE263 pKa = 4.4QYY265 pKa = 9.54RR266 pKa = 11.84TVQADD271 pKa = 3.33SPEE274 pKa = 4.44TYY276 pKa = 10.03KK277 pKa = 10.95AWLRR281 pKa = 11.84RR282 pKa = 11.84VSTISEE288 pKa = 3.8RR289 pKa = 11.84WDD291 pKa = 3.1
MM1 pKa = 6.86SQRR4 pKa = 11.84TRR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 10.08CLTLNNPTDD17 pKa = 3.69AEE19 pKa = 4.28IASLLDD25 pKa = 3.6VQEE28 pKa = 4.52GTLKK32 pKa = 10.64RR33 pKa = 11.84GFIALEE39 pKa = 4.1VGAEE43 pKa = 4.26GTPHH47 pKa = 6.29LQGFVHH53 pKa = 6.96LKK55 pKa = 8.71NAKK58 pKa = 9.21TGTALKK64 pKa = 10.64KK65 pKa = 10.15MLGSDD70 pKa = 2.95RR71 pKa = 11.84WHH73 pKa = 6.36WGKK76 pKa = 11.0ANGTDD81 pKa = 3.96FEE83 pKa = 4.22NWAYY87 pKa = 10.07IMEE90 pKa = 4.52DD91 pKa = 3.25VEE93 pKa = 5.57GKK95 pKa = 9.9VQGDD99 pKa = 4.48LLIQWGDD106 pKa = 3.24KK107 pKa = 9.61PAEE110 pKa = 4.29EE111 pKa = 5.08GEE113 pKa = 3.94PDD115 pKa = 3.19AWDD118 pKa = 4.45SILMMIEE125 pKa = 3.84AGEE128 pKa = 4.2DD129 pKa = 2.97DD130 pKa = 3.69RR131 pKa = 11.84AIVRR135 pKa = 11.84KK136 pKa = 8.39WPSIAIRR143 pKa = 11.84CQSAIAKK150 pKa = 9.36YY151 pKa = 9.65RR152 pKa = 11.84ASFEE156 pKa = 4.16WAEE159 pKa = 3.89CRR161 pKa = 11.84AWRR164 pKa = 11.84EE165 pKa = 3.79VEE167 pKa = 3.89VEE169 pKa = 4.38YY170 pKa = 10.75ISGPTGCGKK179 pKa = 8.2TRR181 pKa = 11.84EE182 pKa = 4.19ALYY185 pKa = 10.97LPDD188 pKa = 3.69GRR190 pKa = 11.84VNTDD194 pKa = 2.31VYY196 pKa = 11.0RR197 pKa = 11.84CTNGKK202 pKa = 9.16HH203 pKa = 5.88PFDD206 pKa = 4.18MYY208 pKa = 10.91DD209 pKa = 3.42GEE211 pKa = 4.42GTIVFEE217 pKa = 4.61EE218 pKa = 4.53FRR220 pKa = 11.84SQYY223 pKa = 7.73TCRR226 pKa = 11.84DD227 pKa = 3.19MLNWVDD233 pKa = 3.92GHH235 pKa = 6.69PLMLPARR242 pKa = 11.84YY243 pKa = 9.24ADD245 pKa = 3.61RR246 pKa = 11.84MAKK249 pKa = 7.73FTKK252 pKa = 10.51VIILSNWRR260 pKa = 11.84FEE262 pKa = 4.15EE263 pKa = 4.4QYY265 pKa = 9.54RR266 pKa = 11.84TVQADD271 pKa = 3.33SPEE274 pKa = 4.44TYY276 pKa = 10.03KK277 pKa = 10.95AWLRR281 pKa = 11.84RR282 pKa = 11.84VSTISEE288 pKa = 3.8RR289 pKa = 11.84WDD291 pKa = 3.1
Molecular weight: 33.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
566 |
275 |
291 |
283.0 |
32.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.834 ± 0.391 | 1.237 ± 0.55 |
5.654 ± 0.813 | 8.481 ± 0.074 |
3.71 ± 0.412 | 7.951 ± 0.26 |
1.59 ± 0.085 | 3.71 ± 0.964 |
4.24 ± 0.61 | 7.244 ± 0.247 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.357 ± 0.176 | 4.594 ± 1.001 |
4.417 ± 0.425 | 3.357 ± 0.176 |
6.36 ± 1.03 | 5.3 ± 0.556 |
6.714 ± 0.352 | 6.007 ± 0.569 |
3.357 ± 0.512 | 3.887 ± 0.071 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
