
Hubei myriapoda virus 8
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Jingchuvirales; Myriaviridae; Myriavirus; Myriavirus myriapedis
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
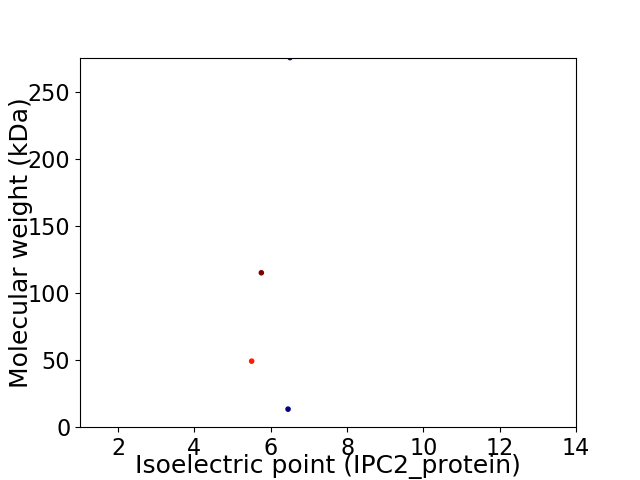
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KN57|A0A1L3KN57_9VIRU Putative glycoprotein OS=Hubei myriapoda virus 8 OX=1922937 PE=4 SV=1
MM1 pKa = 7.59AGAPPFVGLVWQTVLDD17 pKa = 4.14CFKK20 pKa = 10.93DD21 pKa = 3.78LPDD24 pKa = 3.89EE25 pKa = 4.08QKK27 pKa = 10.59LWFGHH32 pKa = 5.6VYY34 pKa = 10.72GGGTGATDD42 pKa = 4.69LIARR46 pKa = 11.84EE47 pKa = 4.15SCLLAMICLKK57 pKa = 10.67YY58 pKa = 10.13RR59 pKa = 11.84VEE61 pKa = 4.1VLRR64 pKa = 11.84QEE66 pKa = 3.93LAIRR70 pKa = 11.84EE71 pKa = 4.14EE72 pKa = 4.33GGLAIKK78 pKa = 9.26TVEE81 pKa = 3.85VLMTEE86 pKa = 4.47NIDD89 pKa = 4.66DD90 pKa = 4.3AMCAPADD97 pKa = 3.96MALHH101 pKa = 7.46WITSALEE108 pKa = 4.3HH109 pKa = 6.27YY110 pKa = 10.34CRR112 pKa = 11.84TPFNVSGVWNNADD125 pKa = 2.86WFEE128 pKa = 4.06YY129 pKa = 10.79DD130 pKa = 3.83HH131 pKa = 7.63IIVAEE136 pKa = 4.28AGAVPAHH143 pKa = 6.29FGGLRR148 pKa = 11.84EE149 pKa = 3.97NHH151 pKa = 6.33GFPGVEE157 pKa = 4.01GNIVRR162 pKa = 11.84ARR164 pKa = 11.84LIRR167 pKa = 11.84DD168 pKa = 3.78LHH170 pKa = 6.69SKK172 pKa = 11.15ADD174 pKa = 3.54VKK176 pKa = 11.14AILVFLLRR184 pKa = 11.84EE185 pKa = 3.91AAVVDD190 pKa = 3.79RR191 pKa = 11.84AEE193 pKa = 4.23RR194 pKa = 11.84GMHH197 pKa = 6.54KK198 pKa = 10.44LFAHH202 pKa = 6.62FFCALSQRR210 pKa = 11.84GNATEE215 pKa = 3.46NWIAKK220 pKa = 8.71RR221 pKa = 11.84QEE223 pKa = 3.88DD224 pKa = 4.92FRR226 pKa = 11.84NQLNDD231 pKa = 3.01QTFVLNSLAMQIIWKK246 pKa = 8.89NWLSKK251 pKa = 10.52IVFPEE256 pKa = 3.97LKK258 pKa = 10.02FVNSLAYY265 pKa = 10.67LEE267 pKa = 5.11ANQLKK272 pKa = 9.04TGQRR276 pKa = 11.84MMLTVEE282 pKa = 4.12HH283 pKa = 6.36SAYY286 pKa = 9.78KK287 pKa = 10.68GLAAYY292 pKa = 9.37GFISSAFNQLPAEE305 pKa = 4.26SFAWAALDD313 pKa = 3.3EE314 pKa = 5.33CIPQNEE320 pKa = 3.74MDD322 pKa = 3.91AYY324 pKa = 10.16EE325 pKa = 4.06IARR328 pKa = 11.84DD329 pKa = 3.65IILEE333 pKa = 4.35GSHH336 pKa = 6.43AMYY339 pKa = 8.29GTRR342 pKa = 11.84EE343 pKa = 3.81VQNRR347 pKa = 11.84LKK349 pKa = 9.93ATRR352 pKa = 11.84YY353 pKa = 8.82PSLAYY358 pKa = 10.3FGVQYY363 pKa = 9.63ATRR366 pKa = 11.84VVRR369 pKa = 11.84NIMANGYY376 pKa = 9.79RR377 pKa = 11.84GVGSNEE383 pKa = 3.22KK384 pKa = 10.47DD385 pKa = 3.47VANRR389 pKa = 11.84ALIDD393 pKa = 4.19EE394 pKa = 5.3IITRR398 pKa = 11.84LQQEE402 pKa = 4.61LGRR405 pKa = 11.84RR406 pKa = 11.84DD407 pKa = 3.84YY408 pKa = 11.12IAGIEE413 pKa = 4.03QNAEE417 pKa = 3.75GRR419 pKa = 11.84NLRR422 pKa = 11.84YY423 pKa = 10.09RR424 pKa = 11.84MLALDD429 pKa = 4.59NPNDD433 pKa = 3.97GNDD436 pKa = 3.07VV437 pKa = 3.39
MM1 pKa = 7.59AGAPPFVGLVWQTVLDD17 pKa = 4.14CFKK20 pKa = 10.93DD21 pKa = 3.78LPDD24 pKa = 3.89EE25 pKa = 4.08QKK27 pKa = 10.59LWFGHH32 pKa = 5.6VYY34 pKa = 10.72GGGTGATDD42 pKa = 4.69LIARR46 pKa = 11.84EE47 pKa = 4.15SCLLAMICLKK57 pKa = 10.67YY58 pKa = 10.13RR59 pKa = 11.84VEE61 pKa = 4.1VLRR64 pKa = 11.84QEE66 pKa = 3.93LAIRR70 pKa = 11.84EE71 pKa = 4.14EE72 pKa = 4.33GGLAIKK78 pKa = 9.26TVEE81 pKa = 3.85VLMTEE86 pKa = 4.47NIDD89 pKa = 4.66DD90 pKa = 4.3AMCAPADD97 pKa = 3.96MALHH101 pKa = 7.46WITSALEE108 pKa = 4.3HH109 pKa = 6.27YY110 pKa = 10.34CRR112 pKa = 11.84TPFNVSGVWNNADD125 pKa = 2.86WFEE128 pKa = 4.06YY129 pKa = 10.79DD130 pKa = 3.83HH131 pKa = 7.63IIVAEE136 pKa = 4.28AGAVPAHH143 pKa = 6.29FGGLRR148 pKa = 11.84EE149 pKa = 3.97NHH151 pKa = 6.33GFPGVEE157 pKa = 4.01GNIVRR162 pKa = 11.84ARR164 pKa = 11.84LIRR167 pKa = 11.84DD168 pKa = 3.78LHH170 pKa = 6.69SKK172 pKa = 11.15ADD174 pKa = 3.54VKK176 pKa = 11.14AILVFLLRR184 pKa = 11.84EE185 pKa = 3.91AAVVDD190 pKa = 3.79RR191 pKa = 11.84AEE193 pKa = 4.23RR194 pKa = 11.84GMHH197 pKa = 6.54KK198 pKa = 10.44LFAHH202 pKa = 6.62FFCALSQRR210 pKa = 11.84GNATEE215 pKa = 3.46NWIAKK220 pKa = 8.71RR221 pKa = 11.84QEE223 pKa = 3.88DD224 pKa = 4.92FRR226 pKa = 11.84NQLNDD231 pKa = 3.01QTFVLNSLAMQIIWKK246 pKa = 8.89NWLSKK251 pKa = 10.52IVFPEE256 pKa = 3.97LKK258 pKa = 10.02FVNSLAYY265 pKa = 10.67LEE267 pKa = 5.11ANQLKK272 pKa = 9.04TGQRR276 pKa = 11.84MMLTVEE282 pKa = 4.12HH283 pKa = 6.36SAYY286 pKa = 9.78KK287 pKa = 10.68GLAAYY292 pKa = 9.37GFISSAFNQLPAEE305 pKa = 4.26SFAWAALDD313 pKa = 3.3EE314 pKa = 5.33CIPQNEE320 pKa = 3.74MDD322 pKa = 3.91AYY324 pKa = 10.16EE325 pKa = 4.06IARR328 pKa = 11.84DD329 pKa = 3.65IILEE333 pKa = 4.35GSHH336 pKa = 6.43AMYY339 pKa = 8.29GTRR342 pKa = 11.84EE343 pKa = 3.81VQNRR347 pKa = 11.84LKK349 pKa = 9.93ATRR352 pKa = 11.84YY353 pKa = 8.82PSLAYY358 pKa = 10.3FGVQYY363 pKa = 9.63ATRR366 pKa = 11.84VVRR369 pKa = 11.84NIMANGYY376 pKa = 9.79RR377 pKa = 11.84GVGSNEE383 pKa = 3.22KK384 pKa = 10.47DD385 pKa = 3.47VANRR389 pKa = 11.84ALIDD393 pKa = 4.19EE394 pKa = 5.3IITRR398 pKa = 11.84LQQEE402 pKa = 4.61LGRR405 pKa = 11.84RR406 pKa = 11.84DD407 pKa = 3.84YY408 pKa = 11.12IAGIEE413 pKa = 4.03QNAEE417 pKa = 3.75GRR419 pKa = 11.84NLRR422 pKa = 11.84YY423 pKa = 10.09RR424 pKa = 11.84MLALDD429 pKa = 4.59NPNDD433 pKa = 3.97GNDD436 pKa = 3.07VV437 pKa = 3.39
Molecular weight: 49.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KN84|A0A1L3KN84_9VIRU Uncharacterized protein OS=Hubei myriapoda virus 8 OX=1922937 PE=4 SV=1
MM1 pKa = 7.47TSFNATGLPTIRR13 pKa = 11.84PGRR16 pKa = 11.84SASVPPSTEE25 pKa = 3.8GAHH28 pKa = 7.2DD29 pKa = 3.55IQKK32 pKa = 10.04PAMFSTMPTAGTLLKK47 pKa = 8.4PTPPYY52 pKa = 9.04MHH54 pKa = 8.16PDD56 pKa = 3.33RR57 pKa = 11.84PLTKK61 pKa = 10.28GDD63 pKa = 3.54FDD65 pKa = 3.71IAMKK69 pKa = 10.6RR70 pKa = 11.84LEE72 pKa = 4.13AQIADD77 pKa = 3.74LTQNVHH83 pKa = 5.12QSIRR87 pKa = 11.84LHH89 pKa = 5.26QQAQGFGTSYY99 pKa = 11.23APPTAPSLYY108 pKa = 9.87PDD110 pKa = 3.46MTEE113 pKa = 4.14QLHH116 pKa = 5.89QGLDD120 pKa = 3.23RR121 pKa = 11.84YY122 pKa = 10.51LL123 pKa = 4.94
MM1 pKa = 7.47TSFNATGLPTIRR13 pKa = 11.84PGRR16 pKa = 11.84SASVPPSTEE25 pKa = 3.8GAHH28 pKa = 7.2DD29 pKa = 3.55IQKK32 pKa = 10.04PAMFSTMPTAGTLLKK47 pKa = 8.4PTPPYY52 pKa = 9.04MHH54 pKa = 8.16PDD56 pKa = 3.33RR57 pKa = 11.84PLTKK61 pKa = 10.28GDD63 pKa = 3.54FDD65 pKa = 3.71IAMKK69 pKa = 10.6RR70 pKa = 11.84LEE72 pKa = 4.13AQIADD77 pKa = 3.74LTQNVHH83 pKa = 5.12QSIRR87 pKa = 11.84LHH89 pKa = 5.26QQAQGFGTSYY99 pKa = 11.23APPTAPSLYY108 pKa = 9.87PDD110 pKa = 3.46MTEE113 pKa = 4.14QLHH116 pKa = 5.89QGLDD120 pKa = 3.23RR121 pKa = 11.84YY122 pKa = 10.51LL123 pKa = 4.94
Molecular weight: 13.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3956 |
123 |
2379 |
989.0 |
113.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.612 ± 1.467 | 1.668 ± 0.128 |
5.207 ± 0.481 | 6.041 ± 0.57 |
4.752 ± 0.198 | 4.424 ± 0.717 |
2.755 ± 0.367 | 8.645 ± 1.083 |
5.485 ± 0.841 | 11.552 ± 0.514 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.705 ± 0.282 | 5.814 ± 0.307 |
3.969 ± 0.463 | 4.146 ± 0.263 |
5.131 ± 0.947 | 6.775 ± 0.89 |
5.688 ± 0.447 | 4.803 ± 0.426 |
1.365 ± 0.139 | 3.463 ± 0.438 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
