
Wenling crustacean virus 13
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Jingchuvirales; Chuviridae; Mivirus; Crustacean mivirus
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
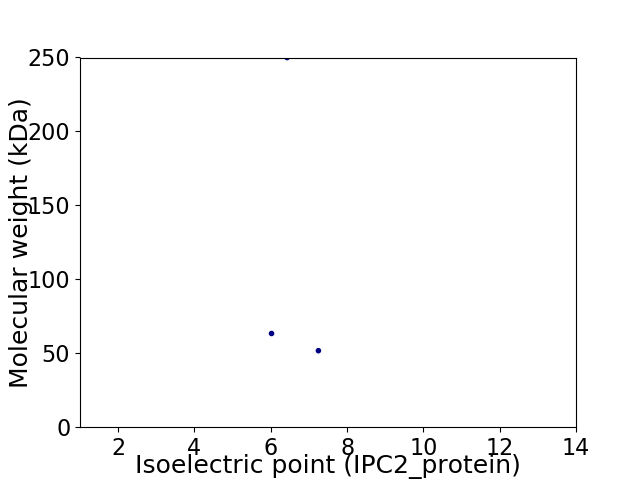
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KNB5|A0A1L3KNB5_9VIRU Putative glycoprotein OS=Wenling crustacean virus 13 OX=1923482 PE=4 SV=1
MM1 pKa = 7.48HH2 pKa = 6.51SHH4 pKa = 5.8VSIVHH9 pKa = 6.49GGHH12 pKa = 5.65EE13 pKa = 4.48AGRR16 pKa = 11.84MTLLPNQCLDD26 pKa = 3.18MHH28 pKa = 7.6KK29 pKa = 11.05NMAYY33 pKa = 8.68QHH35 pKa = 6.56HH36 pKa = 5.81GHH38 pKa = 6.07MVLLDD43 pKa = 4.0GPNTITDD50 pKa = 3.39ATITVAGSVDD60 pKa = 3.31SSGWCEE66 pKa = 3.56GSSFSHH72 pKa = 6.7RR73 pKa = 11.84GSSYY77 pKa = 8.49TSVVVVDD84 pKa = 3.52TLTIKK89 pKa = 10.53VKK91 pKa = 10.65QVDD94 pKa = 4.0ALVQATSGEE103 pKa = 4.1VHH105 pKa = 6.76LPNGVSCLLGSKK117 pKa = 9.98DD118 pKa = 4.33CYY120 pKa = 10.45HH121 pKa = 7.79PIYY124 pKa = 10.75GMAVIDD130 pKa = 3.66EE131 pKa = 4.55TNIGGCGDD139 pKa = 4.14DD140 pKa = 3.31SHH142 pKa = 7.68EE143 pKa = 4.79IIYY146 pKa = 9.15EE147 pKa = 4.13GPIEE151 pKa = 4.17MVSMFGDD158 pKa = 3.2SGEE161 pKa = 4.04DD162 pKa = 2.94EE163 pKa = 4.78RR164 pKa = 11.84YY165 pKa = 9.47FVVEE169 pKa = 3.93TPEE172 pKa = 3.96TVFALKK178 pKa = 10.08RR179 pKa = 11.84GKK181 pKa = 10.07KK182 pKa = 9.31IQVCRR187 pKa = 11.84EE188 pKa = 4.02NAWSTEE194 pKa = 3.88HH195 pKa = 6.96PKK197 pKa = 10.84LIVIPKK203 pKa = 8.78PEE205 pKa = 3.92WGFKK209 pKa = 6.94FTKK212 pKa = 9.93RR213 pKa = 11.84ISGASSVHH221 pKa = 6.77LIPYY225 pKa = 8.99VNSKK229 pKa = 9.83IQFLDD234 pKa = 3.53LKK236 pKa = 10.56VGQNMVRR243 pKa = 11.84TKK245 pKa = 10.84VEE247 pKa = 4.06LLEE250 pKa = 4.34QICEE254 pKa = 3.94NSRR257 pKa = 11.84KK258 pKa = 9.32IVEE261 pKa = 4.15NRR263 pKa = 11.84LMLAKK268 pKa = 10.57LFPHH272 pKa = 5.99QVASLHH278 pKa = 5.21YY279 pKa = 9.42NKK281 pKa = 9.27PGYY284 pKa = 8.53MGRR287 pKa = 11.84VSGEE291 pKa = 3.52ALHH294 pKa = 7.13IIKK297 pKa = 10.14CKK299 pKa = 9.25PVSVSGRR306 pKa = 11.84KK307 pKa = 9.32AEE309 pKa = 4.07GCYY312 pKa = 9.93QGIPVTHH319 pKa = 6.86KK320 pKa = 10.28NEE322 pKa = 3.93SWFLLPITRR331 pKa = 11.84ILSRR335 pKa = 11.84NAVEE339 pKa = 4.65TTCSRR344 pKa = 11.84RR345 pKa = 11.84LPNVYY350 pKa = 10.36LFGDD354 pKa = 3.94SWWEE358 pKa = 3.7LGPEE362 pKa = 4.18PRR364 pKa = 11.84PAQPPKK370 pKa = 10.17TMTVGSLLPQWGRR383 pKa = 11.84TRR385 pKa = 11.84TSPLGFGGLYY395 pKa = 9.94PYY397 pKa = 10.41RR398 pKa = 11.84DD399 pKa = 3.2MMAYY403 pKa = 9.04EE404 pKa = 3.93QALFGPVVTDD414 pKa = 2.94TGIAIMTRR422 pKa = 11.84KK423 pKa = 6.05MTGMASHH430 pKa = 7.86DD431 pKa = 3.73EE432 pKa = 4.53TFSSTKK438 pKa = 10.65LFDD441 pKa = 3.61QDD443 pKa = 5.27DD444 pKa = 3.83MGSFQDD450 pKa = 5.66SLIEE454 pKa = 4.48HH455 pKa = 6.3NWGPLAWMGEE465 pKa = 3.88ILAEE469 pKa = 4.15GAAVIVCIVLILLLLKK485 pKa = 10.68LVVRR489 pKa = 11.84VYY491 pKa = 11.0KK492 pKa = 10.63LWRR495 pKa = 11.84VSGAGPWLLWSILSVFTEE513 pKa = 5.08TYY515 pKa = 9.17TAIKK519 pKa = 10.28FITNLDD525 pKa = 4.34KK526 pKa = 10.93IPCSACHH533 pKa = 6.66PCTCHH538 pKa = 6.22TRR540 pKa = 11.84TDD542 pKa = 3.49STQEE546 pKa = 3.4IDD548 pKa = 3.69SEE550 pKa = 4.2NGRR553 pKa = 11.84LYY555 pKa = 10.38PVFSHH560 pKa = 7.33ADD562 pKa = 3.06IAVTNAEE569 pKa = 4.28VAA571 pKa = 3.53
MM1 pKa = 7.48HH2 pKa = 6.51SHH4 pKa = 5.8VSIVHH9 pKa = 6.49GGHH12 pKa = 5.65EE13 pKa = 4.48AGRR16 pKa = 11.84MTLLPNQCLDD26 pKa = 3.18MHH28 pKa = 7.6KK29 pKa = 11.05NMAYY33 pKa = 8.68QHH35 pKa = 6.56HH36 pKa = 5.81GHH38 pKa = 6.07MVLLDD43 pKa = 4.0GPNTITDD50 pKa = 3.39ATITVAGSVDD60 pKa = 3.31SSGWCEE66 pKa = 3.56GSSFSHH72 pKa = 6.7RR73 pKa = 11.84GSSYY77 pKa = 8.49TSVVVVDD84 pKa = 3.52TLTIKK89 pKa = 10.53VKK91 pKa = 10.65QVDD94 pKa = 4.0ALVQATSGEE103 pKa = 4.1VHH105 pKa = 6.76LPNGVSCLLGSKK117 pKa = 9.98DD118 pKa = 4.33CYY120 pKa = 10.45HH121 pKa = 7.79PIYY124 pKa = 10.75GMAVIDD130 pKa = 3.66EE131 pKa = 4.55TNIGGCGDD139 pKa = 4.14DD140 pKa = 3.31SHH142 pKa = 7.68EE143 pKa = 4.79IIYY146 pKa = 9.15EE147 pKa = 4.13GPIEE151 pKa = 4.17MVSMFGDD158 pKa = 3.2SGEE161 pKa = 4.04DD162 pKa = 2.94EE163 pKa = 4.78RR164 pKa = 11.84YY165 pKa = 9.47FVVEE169 pKa = 3.93TPEE172 pKa = 3.96TVFALKK178 pKa = 10.08RR179 pKa = 11.84GKK181 pKa = 10.07KK182 pKa = 9.31IQVCRR187 pKa = 11.84EE188 pKa = 4.02NAWSTEE194 pKa = 3.88HH195 pKa = 6.96PKK197 pKa = 10.84LIVIPKK203 pKa = 8.78PEE205 pKa = 3.92WGFKK209 pKa = 6.94FTKK212 pKa = 9.93RR213 pKa = 11.84ISGASSVHH221 pKa = 6.77LIPYY225 pKa = 8.99VNSKK229 pKa = 9.83IQFLDD234 pKa = 3.53LKK236 pKa = 10.56VGQNMVRR243 pKa = 11.84TKK245 pKa = 10.84VEE247 pKa = 4.06LLEE250 pKa = 4.34QICEE254 pKa = 3.94NSRR257 pKa = 11.84KK258 pKa = 9.32IVEE261 pKa = 4.15NRR263 pKa = 11.84LMLAKK268 pKa = 10.57LFPHH272 pKa = 5.99QVASLHH278 pKa = 5.21YY279 pKa = 9.42NKK281 pKa = 9.27PGYY284 pKa = 8.53MGRR287 pKa = 11.84VSGEE291 pKa = 3.52ALHH294 pKa = 7.13IIKK297 pKa = 10.14CKK299 pKa = 9.25PVSVSGRR306 pKa = 11.84KK307 pKa = 9.32AEE309 pKa = 4.07GCYY312 pKa = 9.93QGIPVTHH319 pKa = 6.86KK320 pKa = 10.28NEE322 pKa = 3.93SWFLLPITRR331 pKa = 11.84ILSRR335 pKa = 11.84NAVEE339 pKa = 4.65TTCSRR344 pKa = 11.84RR345 pKa = 11.84LPNVYY350 pKa = 10.36LFGDD354 pKa = 3.94SWWEE358 pKa = 3.7LGPEE362 pKa = 4.18PRR364 pKa = 11.84PAQPPKK370 pKa = 10.17TMTVGSLLPQWGRR383 pKa = 11.84TRR385 pKa = 11.84TSPLGFGGLYY395 pKa = 9.94PYY397 pKa = 10.41RR398 pKa = 11.84DD399 pKa = 3.2MMAYY403 pKa = 9.04EE404 pKa = 3.93QALFGPVVTDD414 pKa = 2.94TGIAIMTRR422 pKa = 11.84KK423 pKa = 6.05MTGMASHH430 pKa = 7.86DD431 pKa = 3.73EE432 pKa = 4.53TFSSTKK438 pKa = 10.65LFDD441 pKa = 3.61QDD443 pKa = 5.27DD444 pKa = 3.83MGSFQDD450 pKa = 5.66SLIEE454 pKa = 4.48HH455 pKa = 6.3NWGPLAWMGEE465 pKa = 3.88ILAEE469 pKa = 4.15GAAVIVCIVLILLLLKK485 pKa = 10.68LVVRR489 pKa = 11.84VYY491 pKa = 11.0KK492 pKa = 10.63LWRR495 pKa = 11.84VSGAGPWLLWSILSVFTEE513 pKa = 5.08TYY515 pKa = 9.17TAIKK519 pKa = 10.28FITNLDD525 pKa = 4.34KK526 pKa = 10.93IPCSACHH533 pKa = 6.66PCTCHH538 pKa = 6.22TRR540 pKa = 11.84TDD542 pKa = 3.49STQEE546 pKa = 3.4IDD548 pKa = 3.69SEE550 pKa = 4.2NGRR553 pKa = 11.84LYY555 pKa = 10.38PVFSHH560 pKa = 7.33ADD562 pKa = 3.06IAVTNAEE569 pKa = 4.28VAA571 pKa = 3.53
Molecular weight: 63.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KN74|A0A1L3KN74_9VIRU Large structural protein OS=Wenling crustacean virus 13 OX=1923482 PE=4 SV=1
MM1 pKa = 7.96KK2 pKa = 10.15IPQILLKK9 pKa = 10.03HH10 pKa = 5.99PPVLYY15 pKa = 10.29VVVVMAQHH23 pKa = 4.74VHH25 pKa = 5.87LVNGLLDD32 pKa = 3.51KK33 pKa = 11.29AGFWRR38 pKa = 11.84EE39 pKa = 3.71VLKK42 pKa = 11.17QEE44 pKa = 4.2DD45 pKa = 5.1PYY47 pKa = 11.34MKK49 pKa = 10.45LCLRR53 pKa = 11.84HH54 pKa = 6.09CNARR58 pKa = 11.84PWVAPDD64 pKa = 5.22GIRR67 pKa = 11.84KK68 pKa = 8.45WGRR71 pKa = 11.84AMEE74 pKa = 4.38VAWLLSTRR82 pKa = 11.84IVATLRR88 pKa = 11.84VTTPHH93 pKa = 6.9SILVWTMMEE102 pKa = 3.85TLAPQVHH109 pKa = 6.91DD110 pKa = 3.46EE111 pKa = 4.27RR112 pKa = 11.84YY113 pKa = 9.51IPRR116 pKa = 11.84LEE118 pKa = 4.1ANALIVRR125 pKa = 11.84SVYY128 pKa = 10.61NDD130 pKa = 3.17SSFQSLVSNYY140 pKa = 8.97TDD142 pKa = 3.81ANLAQDD148 pKa = 4.27LHH150 pKa = 7.35DD151 pKa = 5.31LSQDD155 pKa = 3.34PDD157 pKa = 3.74FEE159 pKa = 5.3RR160 pKa = 11.84PANDD164 pKa = 3.86DD165 pKa = 3.72GVPTTIAIQFSAVANKK181 pKa = 10.1SAEE184 pKa = 3.95GAMAILWVLAKK195 pKa = 10.43RR196 pKa = 11.84SVNLHH201 pKa = 5.06TRR203 pKa = 11.84PAQVLVHH210 pKa = 5.94TLCALAKK217 pKa = 10.15QGNATQAFVEE227 pKa = 5.05KK228 pKa = 9.19ITQGFVTDD236 pKa = 3.36TGKK239 pKa = 9.74PLNLDD244 pKa = 3.3AEE246 pKa = 4.95GIEE249 pKa = 4.28ACWRR253 pKa = 11.84LIVHH257 pKa = 6.67HH258 pKa = 7.02VDD260 pKa = 3.5DD261 pKa = 5.35SNVEE265 pKa = 4.31AIVSDD270 pKa = 3.78WLAKK274 pKa = 10.29LPHH277 pKa = 6.09EE278 pKa = 4.43ALRR281 pKa = 11.84LRR283 pKa = 11.84ITLQQIPGEE292 pKa = 4.24GLTCITSIIKK302 pKa = 10.36AIQDD306 pKa = 3.38FPGFNWTWVYY316 pKa = 11.5KK317 pKa = 10.25NFPGEE322 pKa = 3.72MTAAGTASIAITNRR336 pKa = 11.84GFYY339 pKa = 10.23GYY341 pKa = 10.55KK342 pKa = 9.73KK343 pKa = 10.44DD344 pKa = 3.76LSLVKK349 pKa = 10.1AANFRR354 pKa = 11.84TVGYY358 pKa = 9.74IAQQLLVRR366 pKa = 11.84AGGEE370 pKa = 4.33GPLRR374 pKa = 11.84AAKK377 pKa = 10.43CFTRR381 pKa = 11.84TPKK384 pKa = 10.61LKK386 pKa = 10.76ALIDD390 pKa = 3.6QMIDD394 pKa = 3.32DD395 pKa = 5.28FIINAVTIPVNPGDD409 pKa = 4.27EE410 pKa = 4.34VPAGIWASIMGYY422 pKa = 8.64SVPPAPQPVLMAGPQPGGGHH442 pKa = 6.9GGGQLPPGGPGPGAPPAPPQPQAGPNVQGGNRR474 pKa = 11.84PP475 pKa = 3.35
MM1 pKa = 7.96KK2 pKa = 10.15IPQILLKK9 pKa = 10.03HH10 pKa = 5.99PPVLYY15 pKa = 10.29VVVVMAQHH23 pKa = 4.74VHH25 pKa = 5.87LVNGLLDD32 pKa = 3.51KK33 pKa = 11.29AGFWRR38 pKa = 11.84EE39 pKa = 3.71VLKK42 pKa = 11.17QEE44 pKa = 4.2DD45 pKa = 5.1PYY47 pKa = 11.34MKK49 pKa = 10.45LCLRR53 pKa = 11.84HH54 pKa = 6.09CNARR58 pKa = 11.84PWVAPDD64 pKa = 5.22GIRR67 pKa = 11.84KK68 pKa = 8.45WGRR71 pKa = 11.84AMEE74 pKa = 4.38VAWLLSTRR82 pKa = 11.84IVATLRR88 pKa = 11.84VTTPHH93 pKa = 6.9SILVWTMMEE102 pKa = 3.85TLAPQVHH109 pKa = 6.91DD110 pKa = 3.46EE111 pKa = 4.27RR112 pKa = 11.84YY113 pKa = 9.51IPRR116 pKa = 11.84LEE118 pKa = 4.1ANALIVRR125 pKa = 11.84SVYY128 pKa = 10.61NDD130 pKa = 3.17SSFQSLVSNYY140 pKa = 8.97TDD142 pKa = 3.81ANLAQDD148 pKa = 4.27LHH150 pKa = 7.35DD151 pKa = 5.31LSQDD155 pKa = 3.34PDD157 pKa = 3.74FEE159 pKa = 5.3RR160 pKa = 11.84PANDD164 pKa = 3.86DD165 pKa = 3.72GVPTTIAIQFSAVANKK181 pKa = 10.1SAEE184 pKa = 3.95GAMAILWVLAKK195 pKa = 10.43RR196 pKa = 11.84SVNLHH201 pKa = 5.06TRR203 pKa = 11.84PAQVLVHH210 pKa = 5.94TLCALAKK217 pKa = 10.15QGNATQAFVEE227 pKa = 5.05KK228 pKa = 9.19ITQGFVTDD236 pKa = 3.36TGKK239 pKa = 9.74PLNLDD244 pKa = 3.3AEE246 pKa = 4.95GIEE249 pKa = 4.28ACWRR253 pKa = 11.84LIVHH257 pKa = 6.67HH258 pKa = 7.02VDD260 pKa = 3.5DD261 pKa = 5.35SNVEE265 pKa = 4.31AIVSDD270 pKa = 3.78WLAKK274 pKa = 10.29LPHH277 pKa = 6.09EE278 pKa = 4.43ALRR281 pKa = 11.84LRR283 pKa = 11.84ITLQQIPGEE292 pKa = 4.24GLTCITSIIKK302 pKa = 10.36AIQDD306 pKa = 3.38FPGFNWTWVYY316 pKa = 11.5KK317 pKa = 10.25NFPGEE322 pKa = 3.72MTAAGTASIAITNRR336 pKa = 11.84GFYY339 pKa = 10.23GYY341 pKa = 10.55KK342 pKa = 9.73KK343 pKa = 10.44DD344 pKa = 3.76LSLVKK349 pKa = 10.1AANFRR354 pKa = 11.84TVGYY358 pKa = 9.74IAQQLLVRR366 pKa = 11.84AGGEE370 pKa = 4.33GPLRR374 pKa = 11.84AAKK377 pKa = 10.43CFTRR381 pKa = 11.84TPKK384 pKa = 10.61LKK386 pKa = 10.76ALIDD390 pKa = 3.6QMIDD394 pKa = 3.32DD395 pKa = 5.28FIINAVTIPVNPGDD409 pKa = 4.27EE410 pKa = 4.34VPAGIWASIMGYY422 pKa = 8.64SVPPAPQPVLMAGPQPGGGHH442 pKa = 6.9GGGQLPPGGPGPGAPPAPPQPQAGPNVQGGNRR474 pKa = 11.84PP475 pKa = 3.35
Molecular weight: 51.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3224 |
475 |
2178 |
1074.7 |
121.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.669 ± 1.071 | 2.233 ± 0.272 |
4.932 ± 0.195 | 6.234 ± 0.84 |
3.226 ± 0.153 | 5.552 ± 1.264 |
3.35 ± 0.239 | 6.328 ± 0.009 |
4.901 ± 0.202 | 10.205 ± 0.682 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.419 ± 0.294 | 3.66 ± 0.192 |
5.18 ± 0.958 | 3.412 ± 0.457 |
5.924 ± 0.783 | 6.886 ± 0.859 |
5.955 ± 0.23 | 7.444 ± 0.447 |
2.171 ± 0.041 | 3.319 ± 0.407 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
