
Marixanthomonas sp. HN-E44
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Marixanthomonas; unclassified Marixanthomonas
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
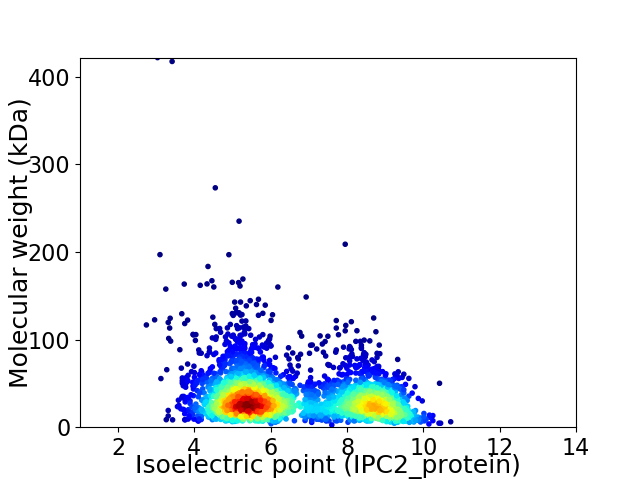
Virtual 2D-PAGE plot for 3068 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U0HX65|A0A2U0HX65_9FLAO Uncharacterized protein OS=Marixanthomonas sp. HN-E44 OX=2174845 GN=DDV96_13620 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 9.05KK3 pKa = 8.31TLLLLITLCCGFCYY17 pKa = 9.6GQNYY21 pKa = 8.91NVSIYY26 pKa = 8.92QQIADD31 pKa = 3.76NKK33 pKa = 9.95GGFNAAIDD41 pKa = 3.59NADD44 pKa = 2.93WFGYY48 pKa = 8.77SVEE51 pKa = 5.08SIGDD55 pKa = 3.77LNGDD59 pKa = 3.77GIEE62 pKa = 4.13DD63 pKa = 3.84VVVGSLKK70 pKa = 10.92DD71 pKa = 3.48DD72 pKa = 4.08DD73 pKa = 5.2GGFNRR78 pKa = 11.84GALYY82 pKa = 10.94VLFLNRR88 pKa = 11.84DD89 pKa = 3.77GEE91 pKa = 4.17VDD93 pKa = 3.76SYY95 pKa = 11.81QKK97 pKa = 10.67ISDD100 pKa = 4.03TEE102 pKa = 4.25GGFQGVLDD110 pKa = 4.27DD111 pKa = 3.86WDD113 pKa = 3.82IFGSSISYY121 pKa = 10.56LGDD124 pKa = 3.18INNDD128 pKa = 2.73GFTEE132 pKa = 3.95IGVGAEE138 pKa = 3.81YY139 pKa = 10.81DD140 pKa = 3.37GDD142 pKa = 3.45GGYY145 pKa = 8.17WHH147 pKa = 6.88GAVWILSLDD156 pKa = 3.61SNGIVQSYY164 pKa = 10.66SKK166 pKa = 10.68ISDD169 pKa = 3.68TEE171 pKa = 4.28GGFLAPLGDD180 pKa = 3.62EE181 pKa = 5.02DD182 pKa = 5.76VFGTDD187 pKa = 2.94IASLGDD193 pKa = 3.71LNGDD197 pKa = 3.58GFGDD201 pKa = 3.38IVVSARR207 pKa = 11.84RR208 pKa = 11.84DD209 pKa = 3.38PDD211 pKa = 3.64GGSDD215 pKa = 3.18RR216 pKa = 11.84GAVYY220 pKa = 10.44ILFLNNDD227 pKa = 3.75LSVNNYY233 pKa = 8.34QKK235 pKa = 10.52ISQTQGNFQGYY246 pKa = 9.68LDD248 pKa = 4.34PGDD251 pKa = 3.93YY252 pKa = 10.8FGGAVANIGDD262 pKa = 3.89IDD264 pKa = 4.88LDD266 pKa = 4.09GVVDD270 pKa = 3.9IAVGAYY276 pKa = 9.72RR277 pKa = 11.84DD278 pKa = 3.93DD279 pKa = 5.58DD280 pKa = 4.26GGIDD284 pKa = 3.38QGAVYY289 pKa = 10.81VLFLNSDD296 pKa = 3.92GSVKK300 pKa = 10.39DD301 pKa = 3.59YY302 pKa = 11.53QKK304 pKa = 10.84ISDD307 pKa = 3.98VYY309 pKa = 11.01GGLEE313 pKa = 3.9TTFNNGMFFGTSIDD327 pKa = 3.48LAYY330 pKa = 10.48DD331 pKa = 3.23INDD334 pKa = 3.46NGLTEE339 pKa = 4.83IIVGAKK345 pKa = 10.16GYY347 pKa = 9.44WNEE350 pKa = 3.93EE351 pKa = 3.68NSEE354 pKa = 3.72IGAFYY359 pKa = 10.51ILEE362 pKa = 4.49LNNNGTVNNSVRR374 pKa = 11.84YY375 pKa = 7.54TKK377 pKa = 10.89GEE379 pKa = 3.85QFFDD383 pKa = 3.67GDD385 pKa = 4.23VVPEE389 pKa = 4.06SSFGFSVSYY398 pKa = 10.93NRR400 pKa = 11.84GLSTNDD406 pKa = 3.57AIVIGAHH413 pKa = 4.59TTGDD417 pKa = 3.07INGFEE422 pKa = 4.44NGSLWILQLGKK433 pKa = 10.05ILSIDD438 pKa = 4.7DD439 pKa = 3.21IDD441 pKa = 4.55GNNEE445 pKa = 3.85QILVYY450 pKa = 9.93PNPTNNSFSLHH461 pKa = 6.62EE462 pKa = 4.81IDD464 pKa = 6.35DD465 pKa = 3.58ILMIQIFDD473 pKa = 3.82SQGKK477 pKa = 9.48EE478 pKa = 3.49IISFKK483 pKa = 10.53DD484 pKa = 2.98IKK486 pKa = 11.2SNNFDD491 pKa = 3.4VSFLPVGVYY500 pKa = 10.18VILIKK505 pKa = 11.01SNNEE509 pKa = 4.18DD510 pKa = 2.91ISSYY514 pKa = 11.46KK515 pKa = 10.35LIKK518 pKa = 10.2NN519 pKa = 3.64
MM1 pKa = 7.67KK2 pKa = 9.05KK3 pKa = 8.31TLLLLITLCCGFCYY17 pKa = 9.6GQNYY21 pKa = 8.91NVSIYY26 pKa = 8.92QQIADD31 pKa = 3.76NKK33 pKa = 9.95GGFNAAIDD41 pKa = 3.59NADD44 pKa = 2.93WFGYY48 pKa = 8.77SVEE51 pKa = 5.08SIGDD55 pKa = 3.77LNGDD59 pKa = 3.77GIEE62 pKa = 4.13DD63 pKa = 3.84VVVGSLKK70 pKa = 10.92DD71 pKa = 3.48DD72 pKa = 4.08DD73 pKa = 5.2GGFNRR78 pKa = 11.84GALYY82 pKa = 10.94VLFLNRR88 pKa = 11.84DD89 pKa = 3.77GEE91 pKa = 4.17VDD93 pKa = 3.76SYY95 pKa = 11.81QKK97 pKa = 10.67ISDD100 pKa = 4.03TEE102 pKa = 4.25GGFQGVLDD110 pKa = 4.27DD111 pKa = 3.86WDD113 pKa = 3.82IFGSSISYY121 pKa = 10.56LGDD124 pKa = 3.18INNDD128 pKa = 2.73GFTEE132 pKa = 3.95IGVGAEE138 pKa = 3.81YY139 pKa = 10.81DD140 pKa = 3.37GDD142 pKa = 3.45GGYY145 pKa = 8.17WHH147 pKa = 6.88GAVWILSLDD156 pKa = 3.61SNGIVQSYY164 pKa = 10.66SKK166 pKa = 10.68ISDD169 pKa = 3.68TEE171 pKa = 4.28GGFLAPLGDD180 pKa = 3.62EE181 pKa = 5.02DD182 pKa = 5.76VFGTDD187 pKa = 2.94IASLGDD193 pKa = 3.71LNGDD197 pKa = 3.58GFGDD201 pKa = 3.38IVVSARR207 pKa = 11.84RR208 pKa = 11.84DD209 pKa = 3.38PDD211 pKa = 3.64GGSDD215 pKa = 3.18RR216 pKa = 11.84GAVYY220 pKa = 10.44ILFLNNDD227 pKa = 3.75LSVNNYY233 pKa = 8.34QKK235 pKa = 10.52ISQTQGNFQGYY246 pKa = 9.68LDD248 pKa = 4.34PGDD251 pKa = 3.93YY252 pKa = 10.8FGGAVANIGDD262 pKa = 3.89IDD264 pKa = 4.88LDD266 pKa = 4.09GVVDD270 pKa = 3.9IAVGAYY276 pKa = 9.72RR277 pKa = 11.84DD278 pKa = 3.93DD279 pKa = 5.58DD280 pKa = 4.26GGIDD284 pKa = 3.38QGAVYY289 pKa = 10.81VLFLNSDD296 pKa = 3.92GSVKK300 pKa = 10.39DD301 pKa = 3.59YY302 pKa = 11.53QKK304 pKa = 10.84ISDD307 pKa = 3.98VYY309 pKa = 11.01GGLEE313 pKa = 3.9TTFNNGMFFGTSIDD327 pKa = 3.48LAYY330 pKa = 10.48DD331 pKa = 3.23INDD334 pKa = 3.46NGLTEE339 pKa = 4.83IIVGAKK345 pKa = 10.16GYY347 pKa = 9.44WNEE350 pKa = 3.93EE351 pKa = 3.68NSEE354 pKa = 3.72IGAFYY359 pKa = 10.51ILEE362 pKa = 4.49LNNNGTVNNSVRR374 pKa = 11.84YY375 pKa = 7.54TKK377 pKa = 10.89GEE379 pKa = 3.85QFFDD383 pKa = 3.67GDD385 pKa = 4.23VVPEE389 pKa = 4.06SSFGFSVSYY398 pKa = 10.93NRR400 pKa = 11.84GLSTNDD406 pKa = 3.57AIVIGAHH413 pKa = 4.59TTGDD417 pKa = 3.07INGFEE422 pKa = 4.44NGSLWILQLGKK433 pKa = 10.05ILSIDD438 pKa = 4.7DD439 pKa = 3.21IDD441 pKa = 4.55GNNEE445 pKa = 3.85QILVYY450 pKa = 9.93PNPTNNSFSLHH461 pKa = 6.62EE462 pKa = 4.81IDD464 pKa = 6.35DD465 pKa = 3.58ILMIQIFDD473 pKa = 3.82SQGKK477 pKa = 9.48EE478 pKa = 3.49IISFKK483 pKa = 10.53DD484 pKa = 2.98IKK486 pKa = 11.2SNNFDD491 pKa = 3.4VSFLPVGVYY500 pKa = 10.18VILIKK505 pKa = 11.01SNNEE509 pKa = 4.18DD510 pKa = 2.91ISSYY514 pKa = 11.46KK515 pKa = 10.35LIKK518 pKa = 10.2NN519 pKa = 3.64
Molecular weight: 56.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U0I0V5|A0A2U0I0V5_9FLAO DUF4168 domain-containing protein OS=Marixanthomonas sp. HN-E44 OX=2174845 GN=DDV96_08880 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.18RR21 pKa = 11.84MASVNGRR28 pKa = 11.84KK29 pKa = 9.17VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.02GRR39 pKa = 11.84KK40 pKa = 8.12KK41 pKa = 10.62LSVSSEE47 pKa = 4.03TRR49 pKa = 11.84HH50 pKa = 5.96KK51 pKa = 10.68KK52 pKa = 9.8
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.18RR21 pKa = 11.84MASVNGRR28 pKa = 11.84KK29 pKa = 9.17VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.02GRR39 pKa = 11.84KK40 pKa = 8.12KK41 pKa = 10.62LSVSSEE47 pKa = 4.03TRR49 pKa = 11.84HH50 pKa = 5.96KK51 pKa = 10.68KK52 pKa = 9.8
Molecular weight: 6.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1006229 |
24 |
4048 |
328.0 |
36.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.701 ± 0.048 | 0.726 ± 0.018 |
5.605 ± 0.039 | 6.92 ± 0.045 |
5.107 ± 0.038 | 6.47 ± 0.051 |
1.797 ± 0.021 | 7.321 ± 0.045 |
7.775 ± 0.063 | 9.221 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.19 ± 0.028 | 5.882 ± 0.046 |
3.555 ± 0.024 | 3.52 ± 0.027 |
3.522 ± 0.037 | 6.109 ± 0.033 |
6.143 ± 0.067 | 6.434 ± 0.037 |
1.01 ± 0.017 | 3.992 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
