
Halobacteriovorax marinus (strain ATCC BAA-682 / DSM 15412 / SJ) (Bacteriovorax marinus)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Oligoflexia; Bacteriovoracales; Halobacteriovoraceae; Halobacteriovorax; Halobacteriovorax marinus
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
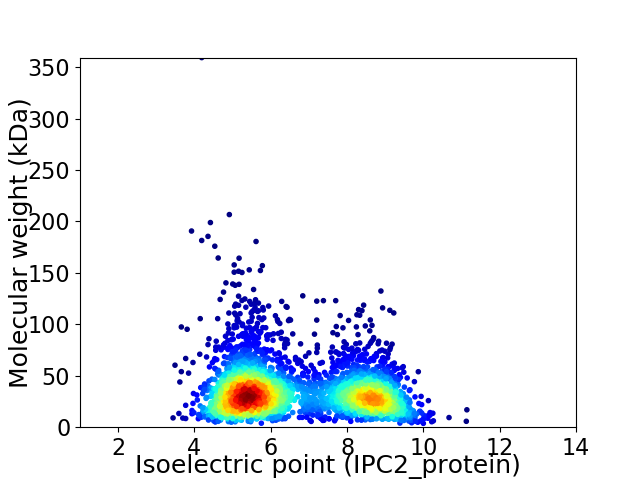
Virtual 2D-PAGE plot for 3230 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
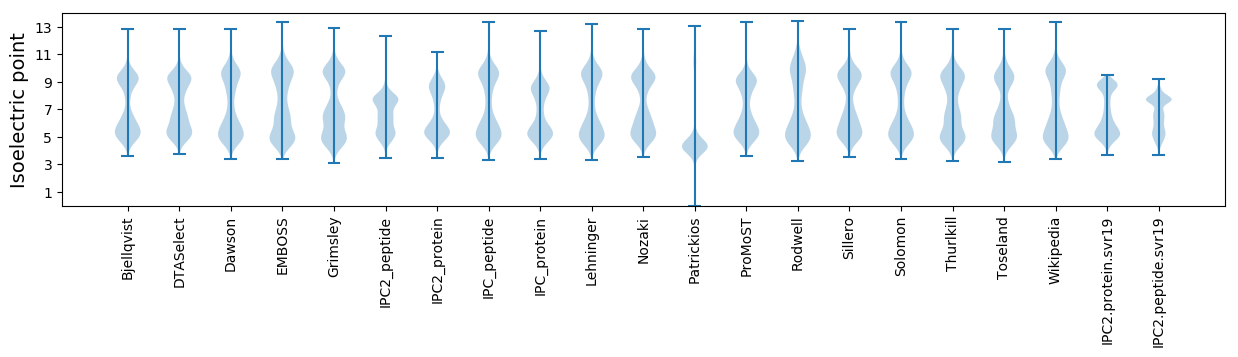
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E1X637|E1X637_HALMS Putative integral membrane zinc-metalloprotease OS=Halobacteriovorax marinus (strain ATCC BAA-682 / DSM 15412 / SJ) OX=862908 GN=BMS_2595 PE=3 SV=1
MM1 pKa = 6.93MVWNFRR7 pKa = 11.84KK8 pKa = 10.14ALTLMSLLTLFAACNSAKK26 pKa = 10.39EE27 pKa = 4.25SVEE30 pKa = 4.08VVDD33 pKa = 3.34QGRR36 pKa = 11.84FNRR39 pKa = 11.84PPVSTPDD46 pKa = 4.09DD47 pKa = 3.83PTNDD51 pKa = 4.61DD52 pKa = 3.23ISLVNSLVAITRR64 pKa = 11.84EE65 pKa = 3.72RR66 pKa = 11.84LAPSDD71 pKa = 3.37QAIITFTARR80 pKa = 11.84DD81 pKa = 3.6SDD83 pKa = 3.66GVAINEE89 pKa = 4.36GGLDD93 pKa = 3.31VDD95 pKa = 5.19FSLVGDD101 pKa = 4.2GTSTGTFSATVDD113 pKa = 3.51TGNGVYY119 pKa = 10.42KK120 pKa = 10.25ATLTGIDD127 pKa = 3.73YY128 pKa = 10.8GSKK131 pKa = 8.14NTIRR135 pKa = 11.84VEE137 pKa = 4.27VEE139 pKa = 3.5GSTLIFVQPLQLQVISGDD157 pKa = 3.66YY158 pKa = 10.12YY159 pKa = 11.32RR160 pKa = 11.84EE161 pKa = 3.72IDD163 pKa = 4.32LDD165 pKa = 3.68NSTDD169 pKa = 3.27QDD171 pKa = 3.5EE172 pKa = 4.58FQIEE176 pKa = 4.5VNLTTSNFDD185 pKa = 3.31YY186 pKa = 11.4SNAEE190 pKa = 3.92SAGEE194 pKa = 4.02DD195 pKa = 2.92IRR197 pKa = 11.84FFDD200 pKa = 4.89EE201 pKa = 5.81DD202 pKa = 4.32FNEE205 pKa = 3.72QDD207 pKa = 3.06FWIEE211 pKa = 3.71NWDD214 pKa = 3.47NTGDD218 pKa = 3.57SKK220 pKa = 11.2IWVKK224 pKa = 10.2VQNSGTDD231 pKa = 3.17KK232 pKa = 11.25LLLVYY237 pKa = 10.48GNNSLASASSRR248 pKa = 11.84EE249 pKa = 4.14GVFSYY254 pKa = 10.94DD255 pKa = 3.44VNKK258 pKa = 10.53DD259 pKa = 2.72IYY261 pKa = 11.19YY262 pKa = 10.28EE263 pKa = 4.26LSQVAGPRR271 pKa = 11.84NYY273 pKa = 10.62GISSYY278 pKa = 10.81ISNNNVDD285 pKa = 3.62VLTNAGYY292 pKa = 10.54SSQTISPVAATTYY305 pKa = 10.88SNVISGLIGVDD316 pKa = 3.96GPISGRR322 pKa = 11.84FLTFNDD328 pKa = 4.17GADD331 pKa = 3.48TVAPLSFASTTLAYY345 pKa = 8.8PKK347 pKa = 10.64SRR349 pKa = 11.84GTDD352 pKa = 2.72DD353 pKa = 3.15WDD355 pKa = 3.38IYY357 pKa = 11.12NPNSVTANFTLSNYY371 pKa = 9.98DD372 pKa = 3.03SSGNLVNSNSYY383 pKa = 10.88SLAAGAEE390 pKa = 3.82LHH392 pKa = 6.68IDD394 pKa = 3.47YY395 pKa = 10.56DD396 pKa = 4.05VSKK399 pKa = 10.07MGLIEE404 pKa = 4.36SDD406 pKa = 4.0YY407 pKa = 10.84PLLGLYY413 pKa = 8.46YY414 pKa = 10.36QSNSNDD420 pKa = 3.18AVVMMKK426 pKa = 10.23PSTDD430 pKa = 3.27IIGPAATSGSIAIVEE445 pKa = 4.6DD446 pKa = 3.81GTNGTIYY453 pKa = 8.97YY454 pKa = 10.11TNGTTQAFSGDD465 pKa = 3.6KK466 pKa = 10.85GDD468 pKa = 4.63VIDD471 pKa = 4.64FSGGGSQNSATGLRR485 pKa = 11.84VIATKK490 pKa = 10.5GVSAVSQADD499 pKa = 3.42SDD501 pKa = 4.18GSEE504 pKa = 4.39SASFWPIEE512 pKa = 4.2EE513 pKa = 4.85LNNDD517 pKa = 4.1YY518 pKa = 10.56IVPASSQYY526 pKa = 11.0VVIICPEE533 pKa = 4.12TVNITLTDD541 pKa = 3.57PGGNDD546 pKa = 3.06NSGVCVPAGGNNPGVISFGSATVVSFQDD574 pKa = 3.54GTRR577 pKa = 11.84VSGDD581 pKa = 3.17ANFFMYY587 pKa = 10.61YY588 pKa = 9.97EE589 pKa = 4.4YY590 pKa = 10.88EE591 pKa = 5.4DD592 pKa = 3.99EE593 pKa = 5.85DD594 pKa = 3.9EE595 pKa = 4.6TNVVSWKK602 pKa = 8.11QARR605 pKa = 11.84SYY607 pKa = 11.46SPVPVSTSVGAEE619 pKa = 3.95QSWDD623 pKa = 3.21
MM1 pKa = 6.93MVWNFRR7 pKa = 11.84KK8 pKa = 10.14ALTLMSLLTLFAACNSAKK26 pKa = 10.39EE27 pKa = 4.25SVEE30 pKa = 4.08VVDD33 pKa = 3.34QGRR36 pKa = 11.84FNRR39 pKa = 11.84PPVSTPDD46 pKa = 4.09DD47 pKa = 3.83PTNDD51 pKa = 4.61DD52 pKa = 3.23ISLVNSLVAITRR64 pKa = 11.84EE65 pKa = 3.72RR66 pKa = 11.84LAPSDD71 pKa = 3.37QAIITFTARR80 pKa = 11.84DD81 pKa = 3.6SDD83 pKa = 3.66GVAINEE89 pKa = 4.36GGLDD93 pKa = 3.31VDD95 pKa = 5.19FSLVGDD101 pKa = 4.2GTSTGTFSATVDD113 pKa = 3.51TGNGVYY119 pKa = 10.42KK120 pKa = 10.25ATLTGIDD127 pKa = 3.73YY128 pKa = 10.8GSKK131 pKa = 8.14NTIRR135 pKa = 11.84VEE137 pKa = 4.27VEE139 pKa = 3.5GSTLIFVQPLQLQVISGDD157 pKa = 3.66YY158 pKa = 10.12YY159 pKa = 11.32RR160 pKa = 11.84EE161 pKa = 3.72IDD163 pKa = 4.32LDD165 pKa = 3.68NSTDD169 pKa = 3.27QDD171 pKa = 3.5EE172 pKa = 4.58FQIEE176 pKa = 4.5VNLTTSNFDD185 pKa = 3.31YY186 pKa = 11.4SNAEE190 pKa = 3.92SAGEE194 pKa = 4.02DD195 pKa = 2.92IRR197 pKa = 11.84FFDD200 pKa = 4.89EE201 pKa = 5.81DD202 pKa = 4.32FNEE205 pKa = 3.72QDD207 pKa = 3.06FWIEE211 pKa = 3.71NWDD214 pKa = 3.47NTGDD218 pKa = 3.57SKK220 pKa = 11.2IWVKK224 pKa = 10.2VQNSGTDD231 pKa = 3.17KK232 pKa = 11.25LLLVYY237 pKa = 10.48GNNSLASASSRR248 pKa = 11.84EE249 pKa = 4.14GVFSYY254 pKa = 10.94DD255 pKa = 3.44VNKK258 pKa = 10.53DD259 pKa = 2.72IYY261 pKa = 11.19YY262 pKa = 10.28EE263 pKa = 4.26LSQVAGPRR271 pKa = 11.84NYY273 pKa = 10.62GISSYY278 pKa = 10.81ISNNNVDD285 pKa = 3.62VLTNAGYY292 pKa = 10.54SSQTISPVAATTYY305 pKa = 10.88SNVISGLIGVDD316 pKa = 3.96GPISGRR322 pKa = 11.84FLTFNDD328 pKa = 4.17GADD331 pKa = 3.48TVAPLSFASTTLAYY345 pKa = 8.8PKK347 pKa = 10.64SRR349 pKa = 11.84GTDD352 pKa = 2.72DD353 pKa = 3.15WDD355 pKa = 3.38IYY357 pKa = 11.12NPNSVTANFTLSNYY371 pKa = 9.98DD372 pKa = 3.03SSGNLVNSNSYY383 pKa = 10.88SLAAGAEE390 pKa = 3.82LHH392 pKa = 6.68IDD394 pKa = 3.47YY395 pKa = 10.56DD396 pKa = 4.05VSKK399 pKa = 10.07MGLIEE404 pKa = 4.36SDD406 pKa = 4.0YY407 pKa = 10.84PLLGLYY413 pKa = 8.46YY414 pKa = 10.36QSNSNDD420 pKa = 3.18AVVMMKK426 pKa = 10.23PSTDD430 pKa = 3.27IIGPAATSGSIAIVEE445 pKa = 4.6DD446 pKa = 3.81GTNGTIYY453 pKa = 8.97YY454 pKa = 10.11TNGTTQAFSGDD465 pKa = 3.6KK466 pKa = 10.85GDD468 pKa = 4.63VIDD471 pKa = 4.64FSGGGSQNSATGLRR485 pKa = 11.84VIATKK490 pKa = 10.5GVSAVSQADD499 pKa = 3.42SDD501 pKa = 4.18GSEE504 pKa = 4.39SASFWPIEE512 pKa = 4.2EE513 pKa = 4.85LNNDD517 pKa = 4.1YY518 pKa = 10.56IVPASSQYY526 pKa = 11.0VVIICPEE533 pKa = 4.12TVNITLTDD541 pKa = 3.57PGGNDD546 pKa = 3.06NSGVCVPAGGNNPGVISFGSATVVSFQDD574 pKa = 3.54GTRR577 pKa = 11.84VSGDD581 pKa = 3.17ANFFMYY587 pKa = 10.61YY588 pKa = 9.97EE589 pKa = 4.4YY590 pKa = 10.88EE591 pKa = 5.4DD592 pKa = 3.99EE593 pKa = 5.85DD594 pKa = 3.9EE595 pKa = 4.6TNVVSWKK602 pKa = 8.11QARR605 pKa = 11.84SYY607 pKa = 11.46SPVPVSTSVGAEE619 pKa = 3.95QSWDD623 pKa = 3.21
Molecular weight: 66.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E1X1N0|E1X1N0_HALMS Putative integral membrane protein OS=Halobacteriovorax marinus (strain ATCC BAA-682 / DSM 15412 / SJ) OX=862908 GN=BMS_0007 PE=4 SV=1
MM1 pKa = 7.83SKK3 pKa = 9.05RR4 pKa = 11.84TWQPKK9 pKa = 7.64RR10 pKa = 11.84KK11 pKa = 9.41KK12 pKa = 9.57RR13 pKa = 11.84LRR15 pKa = 11.84VHH17 pKa = 6.74GFLKK21 pKa = 10.62RR22 pKa = 11.84MSTAGGKK29 pKa = 9.27NVINARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.01GRR40 pKa = 11.84KK41 pKa = 8.14QLTVSTGSKK50 pKa = 10.24
MM1 pKa = 7.83SKK3 pKa = 9.05RR4 pKa = 11.84TWQPKK9 pKa = 7.64RR10 pKa = 11.84KK11 pKa = 9.41KK12 pKa = 9.57RR13 pKa = 11.84LRR15 pKa = 11.84VHH17 pKa = 6.74GFLKK21 pKa = 10.62RR22 pKa = 11.84MSTAGGKK29 pKa = 9.27NVINARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.01GRR40 pKa = 11.84KK41 pKa = 8.14QLTVSTGSKK50 pKa = 10.24
Molecular weight: 5.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1060232 |
33 |
3358 |
328.2 |
37.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.818 ± 0.043 | 1.105 ± 0.017 |
5.543 ± 0.031 | 7.371 ± 0.052 |
5.237 ± 0.039 | 6.11 ± 0.045 |
1.882 ± 0.022 | 7.527 ± 0.04 |
8.229 ± 0.052 | 9.931 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.238 ± 0.023 | 5.282 ± 0.036 |
3.161 ± 0.025 | 2.986 ± 0.024 |
4.103 ± 0.029 | 8.003 ± 0.049 |
4.983 ± 0.039 | 5.956 ± 0.039 |
0.919 ± 0.014 | 3.615 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
