
Caballeronia mineralivorans PML1(12)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Caballeronia; Caballeronia mineralivorans
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
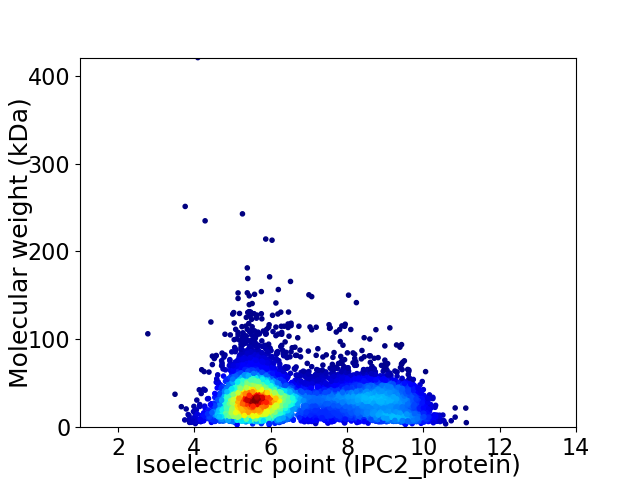
Virtual 2D-PAGE plot for 8182 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0J1D1M2|A0A0J1D1M2_9BURK Sulfate ABC transporter substrate-binding protein OS=Caballeronia mineralivorans PML1(12) OX=908627 GN=EOS_08750 PE=4 SV=1
MM1 pKa = 7.46KK2 pKa = 10.52LILEE6 pKa = 4.41ARR8 pKa = 11.84HH9 pKa = 6.2LFDD12 pKa = 4.66GSVAAVAHH20 pKa = 5.49TTTHH24 pKa = 6.8SDD26 pKa = 3.4TADD29 pKa = 3.68HH30 pKa = 6.18GHH32 pKa = 6.64HH33 pKa = 6.68PVSSDD38 pKa = 2.72LSHH41 pKa = 6.46PHH43 pKa = 5.85SRR45 pKa = 11.84HH46 pKa = 5.55YY47 pKa = 11.19DD48 pKa = 3.54QTQHH52 pKa = 7.25DD53 pKa = 4.32ATLTPTYY60 pKa = 10.86APGDD64 pKa = 3.86IPALAPNPAATEE76 pKa = 3.81ILFIDD81 pKa = 3.75PRR83 pKa = 11.84VANWQALAKK92 pKa = 10.08GVAKK96 pKa = 10.35NVQIVLVDD104 pKa = 3.79PNRR107 pKa = 11.84DD108 pKa = 3.01GLEE111 pKa = 3.95QVTAALKK118 pKa = 10.56GRR120 pKa = 11.84SDD122 pKa = 3.25LTAIQFLTYY131 pKa = 9.59GQSGQIEE138 pKa = 4.97LGNAPITTATLASHH152 pKa = 6.78AGEE155 pKa = 4.05VASWGDD161 pKa = 3.54HH162 pKa = 5.43LAANADD168 pKa = 3.64IEE170 pKa = 4.72FWGCDD175 pKa = 2.93VGQGDD180 pKa = 3.79NGLAFVDD187 pKa = 4.5SVHH190 pKa = 6.81ALTGAQVAASTNATGAAQLGGDD212 pKa = 3.5WTLEE216 pKa = 3.85RR217 pKa = 11.84TTGVLAVGAPFSAASMAAYY236 pKa = 9.9QGVLDD241 pKa = 4.31SPVPTVSFDD250 pKa = 3.48PATVPGDD257 pKa = 3.59VLLGSTFTEE266 pKa = 4.27TVTFANAATNGVGYY280 pKa = 10.73GPFIDD285 pKa = 5.61LYY287 pKa = 9.89VPKK290 pKa = 10.38DD291 pKa = 3.38AAQTATLTGASFLGAGITPEE311 pKa = 4.13AVTLSTTIPGHH322 pKa = 5.41VGVLGALHH330 pKa = 7.17PLALDD335 pKa = 3.48NTGQPLFVAAPPGYY349 pKa = 9.3QANDD353 pKa = 3.03VMYY356 pKa = 10.69VLSLPFGSFTPGQPAAQIQLTFTLDD381 pKa = 3.13NSTEE385 pKa = 3.95LSSMHH390 pKa = 6.75GGQALDD396 pKa = 3.22ITAIGGFQYY405 pKa = 10.94GADD408 pKa = 3.97PLNDD412 pKa = 4.29PGTDD416 pKa = 3.32PSIRR420 pKa = 11.84GTAGTAGVTSAADD433 pKa = 3.56GMAVAASTVSLLDD446 pKa = 3.4VSATTDD452 pKa = 3.18LHH454 pKa = 6.13EE455 pKa = 5.16SEE457 pKa = 4.91TATGPDD463 pKa = 3.36YY464 pKa = 11.0PFNYY468 pKa = 10.15VLTIKK473 pKa = 9.77PAPVTQGDD481 pKa = 4.14PLSNTTFTFDD491 pKa = 4.72LPPEE495 pKa = 4.03VQYY498 pKa = 9.62THH500 pKa = 5.78GTIAFTGPGGATGTATFNPGVAGANGPGGTVTVHH534 pKa = 5.46FTSLGTDD541 pKa = 3.54GADD544 pKa = 2.88TATVVKK550 pKa = 10.35IPVFVPQFDD559 pKa = 4.71ANGNPVLDD567 pKa = 3.94AAGTPRR573 pKa = 11.84DD574 pKa = 3.37ITAATIYY581 pKa = 9.99SYY583 pKa = 10.46TGSWTPTAASLDD595 pKa = 3.78HH596 pKa = 6.91ALGVQTISGDD606 pKa = 3.3SSTNPTSTDD615 pKa = 3.65FVAKK619 pKa = 10.31ALAIQVTDD627 pKa = 4.47DD628 pKa = 3.86APAGNIVPGEE638 pKa = 4.15VVTEE642 pKa = 4.41SIHH645 pKa = 7.48FEE647 pKa = 3.8VSDD650 pKa = 4.25YY651 pKa = 11.73YY652 pKa = 11.6SLNQLKK658 pKa = 10.21IADD661 pKa = 3.87QLGDD665 pKa = 3.94GLSLLSPTEE674 pKa = 3.99LGYY677 pKa = 10.48VAPTLSVTRR686 pKa = 11.84SGVSSAPISFGDD698 pKa = 3.67AGNLVAATVNGQAVAEE714 pKa = 4.39SGSNSAWNYY723 pKa = 8.32TRR725 pKa = 11.84DD726 pKa = 4.43DD727 pKa = 4.31AGTSANPGATSISFGVGALLEE748 pKa = 4.32AQMGAGALASVLQGGGGGATQGVITFVTKK777 pKa = 10.82VLDD780 pKa = 4.15KK781 pKa = 10.41YY782 pKa = 9.56TNTNNEE788 pKa = 3.54DD789 pKa = 3.47SLRR792 pKa = 11.84EE793 pKa = 3.69KK794 pKa = 10.91DD795 pKa = 3.56QITNQVTLAGTSASVVTVDD814 pKa = 3.76NTAQTVGASTGTVADD829 pKa = 3.99NSSVTDD835 pKa = 3.74TVAPGSMVLSVVAVNGQTTDD855 pKa = 3.28LTHH858 pKa = 7.03IEE860 pKa = 4.33PGDD863 pKa = 3.67TVTYY867 pKa = 10.36GLTYY871 pKa = 10.93SLTTGDD877 pKa = 4.39YY878 pKa = 11.37GSLDD882 pKa = 3.44LTANLPLPVFSTTDD896 pKa = 3.39PTSTGSNVTAYY907 pKa = 9.18TADD910 pKa = 3.38NTADD914 pKa = 3.65SFPTAGTYY922 pKa = 10.88KK923 pKa = 10.77LINPLSGEE931 pKa = 4.45SVVSATANGTANSISFNFGNRR952 pKa = 11.84DD953 pKa = 3.43DD954 pKa = 4.05TTNAAGQKK962 pKa = 7.04VTVYY966 pKa = 9.63FSVVASNKK974 pKa = 9.39PFADD978 pKa = 3.98GLPLTSQGDD987 pKa = 3.72SSYY990 pKa = 11.6TNAEE994 pKa = 3.94GQTAATAAIQQVPLEE1009 pKa = 4.26EE1010 pKa = 4.61PEE1012 pKa = 4.3VTVKK1016 pKa = 10.13TGVVSVVGDD1025 pKa = 3.86GSASKK1030 pKa = 9.49GTFTPDD1036 pKa = 2.71TSNPNSAIPWTPQTDD1051 pKa = 3.84VTPSGAAGSPFAPAGTPAGTLFTTVGANGTANPLAADD1088 pKa = 4.23DD1089 pKa = 4.94LNVTGADD1096 pKa = 3.29GGDD1099 pKa = 3.16ITRR1102 pKa = 11.84VVSTVDD1108 pKa = 3.51NEE1110 pKa = 4.17GHH1112 pKa = 6.9ASAFDD1117 pKa = 3.54VTVQGTLPSGYY1128 pKa = 8.64STGNVTNFAIYY1139 pKa = 10.47NSAGAQIDD1147 pKa = 3.81TGVTAAQYY1155 pKa = 9.96FSAGGVKK1162 pKa = 10.52LISTAGTDD1170 pKa = 3.26VGIAAGDD1177 pKa = 3.6RR1178 pKa = 11.84VYY1180 pKa = 11.22VVYY1183 pKa = 10.81DD1184 pKa = 3.58LTLSTTQQTGEE1195 pKa = 4.06TLAAGSSVVNWASVTGGVAAGNGFVSGTGTAALTLGEE1232 pKa = 4.23GSGALSDD1239 pKa = 3.82SATIGISAPSVTKK1252 pKa = 10.02TVTSGSDD1259 pKa = 2.88TALPLGTDD1267 pKa = 3.28NAVVPGEE1274 pKa = 4.32TVTYY1278 pKa = 9.18TVVLTLPQGVTSNGASEE1295 pKa = 4.36LTLTDD1300 pKa = 3.5QLPAGMTFGSISSTSFGTGVTSSNGGTITASSSGSTVTFDD1340 pKa = 4.39LGSSVTNSQVDD1351 pKa = 3.7TNGTITIVYY1360 pKa = 6.94TATVDD1365 pKa = 3.8ATDD1368 pKa = 4.1TPHH1371 pKa = 7.03GGAPTTYY1378 pKa = 10.57SNSAFLNYY1386 pKa = 10.22DD1387 pKa = 3.36SGQTTPVTAANTATVTEE1404 pKa = 4.84HH1405 pKa = 7.07DD1406 pKa = 3.98PQVIEE1411 pKa = 4.91TITAADD1417 pKa = 3.82TTTGVPIATNGTVFSHH1433 pKa = 6.82EE1434 pKa = 4.1GLTYY1438 pKa = 10.42TVTLTNNGTAPANNLADD1455 pKa = 5.26LLDD1458 pKa = 4.34LPAGLTYY1465 pKa = 11.18VNGSLQHH1472 pKa = 5.95VSGGTNSTTSDD1483 pKa = 3.36ANASALNIGLDD1494 pKa = 3.7TLAVGQTATFTFQATVNPNLPAGTSFTVDD1523 pKa = 3.18TPADD1527 pKa = 4.01TTSGTYY1533 pKa = 10.38YY1534 pKa = 10.51SLPGTAQGHH1543 pKa = 6.23KK1544 pKa = 9.91YY1545 pKa = 10.26SDD1547 pKa = 3.79SATDD1551 pKa = 3.23TATIGQITPVLSITGEE1567 pKa = 4.24SNNTDD1572 pKa = 3.3TASTPTQTNTQTSTEE1587 pKa = 4.09AAVGEE1592 pKa = 4.67VVTMQAYY1599 pKa = 9.76VEE1601 pKa = 4.28IPEE1604 pKa = 4.53GSNPTTLNFTMPSGLEE1620 pKa = 3.84YY1621 pKa = 11.06LNDD1624 pKa = 3.78GSAKK1628 pKa = 9.87IAFVSPNGDD1637 pKa = 3.32LVSTDD1642 pKa = 3.47SALSGITQYY1651 pKa = 11.57QDD1653 pKa = 3.12INPGAANYY1661 pKa = 9.27ISPSAITNTGAGTVDD1676 pKa = 3.14VATFRR1681 pKa = 11.84PVDD1684 pKa = 3.82ALPGSVVTATGNSVAIDD1701 pKa = 4.25LGTLSNNDD1709 pKa = 3.02NSSTGNYY1716 pKa = 9.7VLVEE1720 pKa = 4.51FNAVVTNVAANRR1732 pKa = 11.84QGASALQTSFTVAGVTSNDD1751 pKa = 2.7VSVAVEE1757 pKa = 4.41EE1758 pKa = 4.36PAVTVTKK1765 pKa = 9.74TATAVDD1771 pKa = 3.91NTNDD1775 pKa = 3.24TVTYY1779 pKa = 9.95QVTVKK1784 pKa = 9.52NTGLSSAYY1792 pKa = 10.32NVVLDD1797 pKa = 4.7NPAATNEE1804 pKa = 4.14SNIAFVSATGAGTGGTGAGGTTGTDD1829 pKa = 2.94LNYY1832 pKa = 10.85AIGQLGAGGSEE1843 pKa = 4.11VITYY1847 pKa = 6.94TVKK1850 pKa = 10.28VAPGQTVQNDD1860 pKa = 3.48TATATWQSLAGQQTFNGSTAGAVGSVTGPRR1890 pKa = 11.84DD1891 pKa = 3.45FDD1893 pKa = 3.98PAANPPNNYY1902 pKa = 9.41VATAATDD1909 pKa = 2.96IGSAQGRR1916 pKa = 11.84VWQDD1920 pKa = 2.83VGHH1923 pKa = 7.34DD1924 pKa = 3.49KK1925 pKa = 8.59TTFSQTGGSADD1936 pKa = 3.51TALGGITVTATITQPGGTVIHH1957 pKa = 6.35EE1958 pKa = 4.55TTTTASDD1965 pKa = 3.38GTYY1968 pKa = 10.84SFGALPDD1975 pKa = 3.54GTVAISLNGVPANEE1989 pKa = 4.11TLVYY1993 pKa = 10.72DD1994 pKa = 4.06PDD1996 pKa = 4.84GSVSSSPPSASFTSAGDD2013 pKa = 3.26AHH2015 pKa = 6.53NSVDD2019 pKa = 4.92FSFQAPDD2026 pKa = 3.57TAPVIANWSGSTTYY2040 pKa = 11.29AEE2042 pKa = 4.87GGPAMVLSGAGASVSDD2058 pKa = 3.94TQLDD2062 pKa = 3.87ALGGDD2067 pKa = 4.09YY2068 pKa = 11.33SNTTLTLQRR2077 pKa = 11.84YY2078 pKa = 8.33SGGVAAPVNTDD2089 pKa = 3.16VFVGTGLLSLSGSTVTFSGVAVGTFAEE2116 pKa = 4.52ANGKK2120 pKa = 10.13LSITFGAGATAATVKK2135 pKa = 10.69NVLDD2139 pKa = 3.63SLGYY2143 pKa = 10.44SSTDD2147 pKa = 2.79TSTVSTGIQIGATLDD2162 pKa = 3.49DD2163 pKa = 5.13HH2164 pKa = 6.27NTAGAQGTGGNMTSVPAFVTVNEE2187 pKa = 4.21IPAPGGSSATFIEE2200 pKa = 5.09PNDD2203 pKa = 3.73SPAAAVAVAVDD2214 pKa = 3.89STLSVASSDD2223 pKa = 3.62TFAGATVKK2231 pKa = 10.46ISNFKK2236 pKa = 10.17PGEE2239 pKa = 4.18DD2240 pKa = 3.42VLVAGALPSGVTASFDD2256 pKa = 3.57AATGTMTLTGTNLSAATVQAALRR2279 pKa = 11.84GVSYY2283 pKa = 10.82YY2284 pKa = 10.9DD2285 pKa = 4.49SSDD2288 pKa = 3.61TPNTTVRR2295 pKa = 11.84DD2296 pKa = 3.85VTISVMDD2303 pKa = 3.64STTNATTTAAVATIDD2318 pKa = 3.76VVATNDD2324 pKa = 3.49SPLLNGVPVTLNHH2337 pKa = 5.84ATEE2340 pKa = 4.57DD2341 pKa = 3.13AGAPVGAVGTLVSALTGSGNVTDD2364 pKa = 4.65PDD2366 pKa = 3.64GANAHH2371 pKa = 7.08DD2372 pKa = 4.4GTSPGAIGVAITAANTTVGTWWYY2395 pKa = 8.34STNNGATWTEE2405 pKa = 3.91FAGNGTTAVSEE2416 pKa = 4.28ANALHH2421 pKa = 6.68LVADD2425 pKa = 4.61GNTRR2429 pKa = 11.84VYY2431 pKa = 10.74FDD2433 pKa = 5.44PAPDD2437 pKa = 3.55WNGSVPNALTFRR2449 pKa = 11.84AWDD2452 pKa = 3.68QFDD2455 pKa = 3.51GAANGSLSALPAANTLGQGLNTLGSAYY2482 pKa = 10.48SSASQSVDD2490 pKa = 4.31LIGDD2494 pKa = 4.12PVNDD2498 pKa = 3.82APIASGSTTLGSAAEE2513 pKa = 4.09DD2514 pKa = 3.59TAAA2517 pKa = 4.75
MM1 pKa = 7.46KK2 pKa = 10.52LILEE6 pKa = 4.41ARR8 pKa = 11.84HH9 pKa = 6.2LFDD12 pKa = 4.66GSVAAVAHH20 pKa = 5.49TTTHH24 pKa = 6.8SDD26 pKa = 3.4TADD29 pKa = 3.68HH30 pKa = 6.18GHH32 pKa = 6.64HH33 pKa = 6.68PVSSDD38 pKa = 2.72LSHH41 pKa = 6.46PHH43 pKa = 5.85SRR45 pKa = 11.84HH46 pKa = 5.55YY47 pKa = 11.19DD48 pKa = 3.54QTQHH52 pKa = 7.25DD53 pKa = 4.32ATLTPTYY60 pKa = 10.86APGDD64 pKa = 3.86IPALAPNPAATEE76 pKa = 3.81ILFIDD81 pKa = 3.75PRR83 pKa = 11.84VANWQALAKK92 pKa = 10.08GVAKK96 pKa = 10.35NVQIVLVDD104 pKa = 3.79PNRR107 pKa = 11.84DD108 pKa = 3.01GLEE111 pKa = 3.95QVTAALKK118 pKa = 10.56GRR120 pKa = 11.84SDD122 pKa = 3.25LTAIQFLTYY131 pKa = 9.59GQSGQIEE138 pKa = 4.97LGNAPITTATLASHH152 pKa = 6.78AGEE155 pKa = 4.05VASWGDD161 pKa = 3.54HH162 pKa = 5.43LAANADD168 pKa = 3.64IEE170 pKa = 4.72FWGCDD175 pKa = 2.93VGQGDD180 pKa = 3.79NGLAFVDD187 pKa = 4.5SVHH190 pKa = 6.81ALTGAQVAASTNATGAAQLGGDD212 pKa = 3.5WTLEE216 pKa = 3.85RR217 pKa = 11.84TTGVLAVGAPFSAASMAAYY236 pKa = 9.9QGVLDD241 pKa = 4.31SPVPTVSFDD250 pKa = 3.48PATVPGDD257 pKa = 3.59VLLGSTFTEE266 pKa = 4.27TVTFANAATNGVGYY280 pKa = 10.73GPFIDD285 pKa = 5.61LYY287 pKa = 9.89VPKK290 pKa = 10.38DD291 pKa = 3.38AAQTATLTGASFLGAGITPEE311 pKa = 4.13AVTLSTTIPGHH322 pKa = 5.41VGVLGALHH330 pKa = 7.17PLALDD335 pKa = 3.48NTGQPLFVAAPPGYY349 pKa = 9.3QANDD353 pKa = 3.03VMYY356 pKa = 10.69VLSLPFGSFTPGQPAAQIQLTFTLDD381 pKa = 3.13NSTEE385 pKa = 3.95LSSMHH390 pKa = 6.75GGQALDD396 pKa = 3.22ITAIGGFQYY405 pKa = 10.94GADD408 pKa = 3.97PLNDD412 pKa = 4.29PGTDD416 pKa = 3.32PSIRR420 pKa = 11.84GTAGTAGVTSAADD433 pKa = 3.56GMAVAASTVSLLDD446 pKa = 3.4VSATTDD452 pKa = 3.18LHH454 pKa = 6.13EE455 pKa = 5.16SEE457 pKa = 4.91TATGPDD463 pKa = 3.36YY464 pKa = 11.0PFNYY468 pKa = 10.15VLTIKK473 pKa = 9.77PAPVTQGDD481 pKa = 4.14PLSNTTFTFDD491 pKa = 4.72LPPEE495 pKa = 4.03VQYY498 pKa = 9.62THH500 pKa = 5.78GTIAFTGPGGATGTATFNPGVAGANGPGGTVTVHH534 pKa = 5.46FTSLGTDD541 pKa = 3.54GADD544 pKa = 2.88TATVVKK550 pKa = 10.35IPVFVPQFDD559 pKa = 4.71ANGNPVLDD567 pKa = 3.94AAGTPRR573 pKa = 11.84DD574 pKa = 3.37ITAATIYY581 pKa = 9.99SYY583 pKa = 10.46TGSWTPTAASLDD595 pKa = 3.78HH596 pKa = 6.91ALGVQTISGDD606 pKa = 3.3SSTNPTSTDD615 pKa = 3.65FVAKK619 pKa = 10.31ALAIQVTDD627 pKa = 4.47DD628 pKa = 3.86APAGNIVPGEE638 pKa = 4.15VVTEE642 pKa = 4.41SIHH645 pKa = 7.48FEE647 pKa = 3.8VSDD650 pKa = 4.25YY651 pKa = 11.73YY652 pKa = 11.6SLNQLKK658 pKa = 10.21IADD661 pKa = 3.87QLGDD665 pKa = 3.94GLSLLSPTEE674 pKa = 3.99LGYY677 pKa = 10.48VAPTLSVTRR686 pKa = 11.84SGVSSAPISFGDD698 pKa = 3.67AGNLVAATVNGQAVAEE714 pKa = 4.39SGSNSAWNYY723 pKa = 8.32TRR725 pKa = 11.84DD726 pKa = 4.43DD727 pKa = 4.31AGTSANPGATSISFGVGALLEE748 pKa = 4.32AQMGAGALASVLQGGGGGATQGVITFVTKK777 pKa = 10.82VLDD780 pKa = 4.15KK781 pKa = 10.41YY782 pKa = 9.56TNTNNEE788 pKa = 3.54DD789 pKa = 3.47SLRR792 pKa = 11.84EE793 pKa = 3.69KK794 pKa = 10.91DD795 pKa = 3.56QITNQVTLAGTSASVVTVDD814 pKa = 3.76NTAQTVGASTGTVADD829 pKa = 3.99NSSVTDD835 pKa = 3.74TVAPGSMVLSVVAVNGQTTDD855 pKa = 3.28LTHH858 pKa = 7.03IEE860 pKa = 4.33PGDD863 pKa = 3.67TVTYY867 pKa = 10.36GLTYY871 pKa = 10.93SLTTGDD877 pKa = 4.39YY878 pKa = 11.37GSLDD882 pKa = 3.44LTANLPLPVFSTTDD896 pKa = 3.39PTSTGSNVTAYY907 pKa = 9.18TADD910 pKa = 3.38NTADD914 pKa = 3.65SFPTAGTYY922 pKa = 10.88KK923 pKa = 10.77LINPLSGEE931 pKa = 4.45SVVSATANGTANSISFNFGNRR952 pKa = 11.84DD953 pKa = 3.43DD954 pKa = 4.05TTNAAGQKK962 pKa = 7.04VTVYY966 pKa = 9.63FSVVASNKK974 pKa = 9.39PFADD978 pKa = 3.98GLPLTSQGDD987 pKa = 3.72SSYY990 pKa = 11.6TNAEE994 pKa = 3.94GQTAATAAIQQVPLEE1009 pKa = 4.26EE1010 pKa = 4.61PEE1012 pKa = 4.3VTVKK1016 pKa = 10.13TGVVSVVGDD1025 pKa = 3.86GSASKK1030 pKa = 9.49GTFTPDD1036 pKa = 2.71TSNPNSAIPWTPQTDD1051 pKa = 3.84VTPSGAAGSPFAPAGTPAGTLFTTVGANGTANPLAADD1088 pKa = 4.23DD1089 pKa = 4.94LNVTGADD1096 pKa = 3.29GGDD1099 pKa = 3.16ITRR1102 pKa = 11.84VVSTVDD1108 pKa = 3.51NEE1110 pKa = 4.17GHH1112 pKa = 6.9ASAFDD1117 pKa = 3.54VTVQGTLPSGYY1128 pKa = 8.64STGNVTNFAIYY1139 pKa = 10.47NSAGAQIDD1147 pKa = 3.81TGVTAAQYY1155 pKa = 9.96FSAGGVKK1162 pKa = 10.52LISTAGTDD1170 pKa = 3.26VGIAAGDD1177 pKa = 3.6RR1178 pKa = 11.84VYY1180 pKa = 11.22VVYY1183 pKa = 10.81DD1184 pKa = 3.58LTLSTTQQTGEE1195 pKa = 4.06TLAAGSSVVNWASVTGGVAAGNGFVSGTGTAALTLGEE1232 pKa = 4.23GSGALSDD1239 pKa = 3.82SATIGISAPSVTKK1252 pKa = 10.02TVTSGSDD1259 pKa = 2.88TALPLGTDD1267 pKa = 3.28NAVVPGEE1274 pKa = 4.32TVTYY1278 pKa = 9.18TVVLTLPQGVTSNGASEE1295 pKa = 4.36LTLTDD1300 pKa = 3.5QLPAGMTFGSISSTSFGTGVTSSNGGTITASSSGSTVTFDD1340 pKa = 4.39LGSSVTNSQVDD1351 pKa = 3.7TNGTITIVYY1360 pKa = 6.94TATVDD1365 pKa = 3.8ATDD1368 pKa = 4.1TPHH1371 pKa = 7.03GGAPTTYY1378 pKa = 10.57SNSAFLNYY1386 pKa = 10.22DD1387 pKa = 3.36SGQTTPVTAANTATVTEE1404 pKa = 4.84HH1405 pKa = 7.07DD1406 pKa = 3.98PQVIEE1411 pKa = 4.91TITAADD1417 pKa = 3.82TTTGVPIATNGTVFSHH1433 pKa = 6.82EE1434 pKa = 4.1GLTYY1438 pKa = 10.42TVTLTNNGTAPANNLADD1455 pKa = 5.26LLDD1458 pKa = 4.34LPAGLTYY1465 pKa = 11.18VNGSLQHH1472 pKa = 5.95VSGGTNSTTSDD1483 pKa = 3.36ANASALNIGLDD1494 pKa = 3.7TLAVGQTATFTFQATVNPNLPAGTSFTVDD1523 pKa = 3.18TPADD1527 pKa = 4.01TTSGTYY1533 pKa = 10.38YY1534 pKa = 10.51SLPGTAQGHH1543 pKa = 6.23KK1544 pKa = 9.91YY1545 pKa = 10.26SDD1547 pKa = 3.79SATDD1551 pKa = 3.23TATIGQITPVLSITGEE1567 pKa = 4.24SNNTDD1572 pKa = 3.3TASTPTQTNTQTSTEE1587 pKa = 4.09AAVGEE1592 pKa = 4.67VVTMQAYY1599 pKa = 9.76VEE1601 pKa = 4.28IPEE1604 pKa = 4.53GSNPTTLNFTMPSGLEE1620 pKa = 3.84YY1621 pKa = 11.06LNDD1624 pKa = 3.78GSAKK1628 pKa = 9.87IAFVSPNGDD1637 pKa = 3.32LVSTDD1642 pKa = 3.47SALSGITQYY1651 pKa = 11.57QDD1653 pKa = 3.12INPGAANYY1661 pKa = 9.27ISPSAITNTGAGTVDD1676 pKa = 3.14VATFRR1681 pKa = 11.84PVDD1684 pKa = 3.82ALPGSVVTATGNSVAIDD1701 pKa = 4.25LGTLSNNDD1709 pKa = 3.02NSSTGNYY1716 pKa = 9.7VLVEE1720 pKa = 4.51FNAVVTNVAANRR1732 pKa = 11.84QGASALQTSFTVAGVTSNDD1751 pKa = 2.7VSVAVEE1757 pKa = 4.41EE1758 pKa = 4.36PAVTVTKK1765 pKa = 9.74TATAVDD1771 pKa = 3.91NTNDD1775 pKa = 3.24TVTYY1779 pKa = 9.95QVTVKK1784 pKa = 9.52NTGLSSAYY1792 pKa = 10.32NVVLDD1797 pKa = 4.7NPAATNEE1804 pKa = 4.14SNIAFVSATGAGTGGTGAGGTTGTDD1829 pKa = 2.94LNYY1832 pKa = 10.85AIGQLGAGGSEE1843 pKa = 4.11VITYY1847 pKa = 6.94TVKK1850 pKa = 10.28VAPGQTVQNDD1860 pKa = 3.48TATATWQSLAGQQTFNGSTAGAVGSVTGPRR1890 pKa = 11.84DD1891 pKa = 3.45FDD1893 pKa = 3.98PAANPPNNYY1902 pKa = 9.41VATAATDD1909 pKa = 2.96IGSAQGRR1916 pKa = 11.84VWQDD1920 pKa = 2.83VGHH1923 pKa = 7.34DD1924 pKa = 3.49KK1925 pKa = 8.59TTFSQTGGSADD1936 pKa = 3.51TALGGITVTATITQPGGTVIHH1957 pKa = 6.35EE1958 pKa = 4.55TTTTASDD1965 pKa = 3.38GTYY1968 pKa = 10.84SFGALPDD1975 pKa = 3.54GTVAISLNGVPANEE1989 pKa = 4.11TLVYY1993 pKa = 10.72DD1994 pKa = 4.06PDD1996 pKa = 4.84GSVSSSPPSASFTSAGDD2013 pKa = 3.26AHH2015 pKa = 6.53NSVDD2019 pKa = 4.92FSFQAPDD2026 pKa = 3.57TAPVIANWSGSTTYY2040 pKa = 11.29AEE2042 pKa = 4.87GGPAMVLSGAGASVSDD2058 pKa = 3.94TQLDD2062 pKa = 3.87ALGGDD2067 pKa = 4.09YY2068 pKa = 11.33SNTTLTLQRR2077 pKa = 11.84YY2078 pKa = 8.33SGGVAAPVNTDD2089 pKa = 3.16VFVGTGLLSLSGSTVTFSGVAVGTFAEE2116 pKa = 4.52ANGKK2120 pKa = 10.13LSITFGAGATAATVKK2135 pKa = 10.69NVLDD2139 pKa = 3.63SLGYY2143 pKa = 10.44SSTDD2147 pKa = 2.79TSTVSTGIQIGATLDD2162 pKa = 3.49DD2163 pKa = 5.13HH2164 pKa = 6.27NTAGAQGTGGNMTSVPAFVTVNEE2187 pKa = 4.21IPAPGGSSATFIEE2200 pKa = 5.09PNDD2203 pKa = 3.73SPAAAVAVAVDD2214 pKa = 3.89STLSVASSDD2223 pKa = 3.62TFAGATVKK2231 pKa = 10.46ISNFKK2236 pKa = 10.17PGEE2239 pKa = 4.18DD2240 pKa = 3.42VLVAGALPSGVTASFDD2256 pKa = 3.57AATGTMTLTGTNLSAATVQAALRR2279 pKa = 11.84GVSYY2283 pKa = 10.82YY2284 pKa = 10.9DD2285 pKa = 4.49SSDD2288 pKa = 3.61TPNTTVRR2295 pKa = 11.84DD2296 pKa = 3.85VTISVMDD2303 pKa = 3.64STTNATTTAAVATIDD2318 pKa = 3.76VVATNDD2324 pKa = 3.49SPLLNGVPVTLNHH2337 pKa = 5.84ATEE2340 pKa = 4.57DD2341 pKa = 3.13AGAPVGAVGTLVSALTGSGNVTDD2364 pKa = 4.65PDD2366 pKa = 3.64GANAHH2371 pKa = 7.08DD2372 pKa = 4.4GTSPGAIGVAITAANTTVGTWWYY2395 pKa = 8.34STNNGATWTEE2405 pKa = 3.91FAGNGTTAVSEE2416 pKa = 4.28ANALHH2421 pKa = 6.68LVADD2425 pKa = 4.61GNTRR2429 pKa = 11.84VYY2431 pKa = 10.74FDD2433 pKa = 5.44PAPDD2437 pKa = 3.55WNGSVPNALTFRR2449 pKa = 11.84AWDD2452 pKa = 3.68QFDD2455 pKa = 3.51GAANGSLSALPAANTLGQGLNTLGSAYY2482 pKa = 10.48SSASQSVDD2490 pKa = 4.31LIGDD2494 pKa = 4.12PVNDD2498 pKa = 3.82APIASGSTTLGSAAEE2513 pKa = 4.09DD2514 pKa = 3.59TAAA2517 pKa = 4.75
Molecular weight: 251.29 kDa
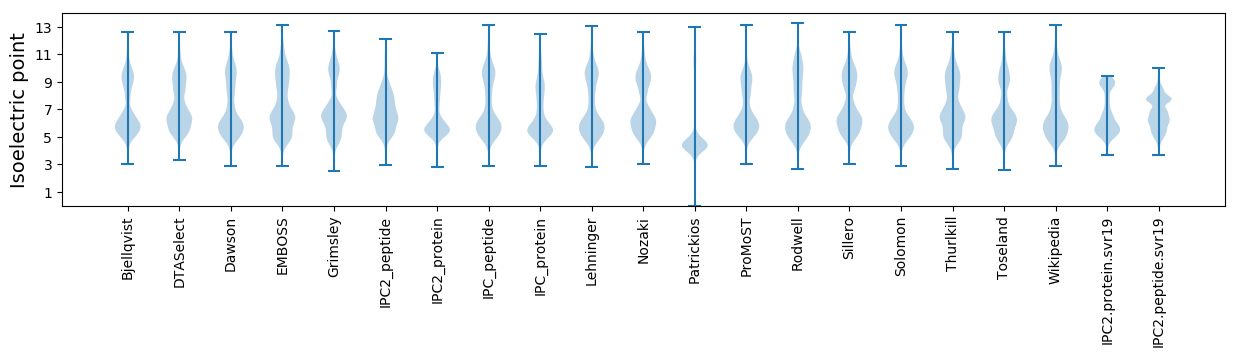
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0J1CR47|A0A0J1CR47_9BURK DEAD/DEAH box helicase OS=Caballeronia mineralivorans PML1(12) OX=908627 GN=EOS_27170 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.06QPSVTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.0RR14 pKa = 11.84THH16 pKa = 5.76GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.74TAGGRR28 pKa = 11.84KK29 pKa = 9.04VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.06QPSVTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.0RR14 pKa = 11.84THH16 pKa = 5.76GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.74TAGGRR28 pKa = 11.84KK29 pKa = 9.04VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2499026 |
24 |
4234 |
305.4 |
33.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.116 ± 0.031 | 0.906 ± 0.008 |
5.382 ± 0.022 | 5.108 ± 0.026 |
3.825 ± 0.017 | 8.114 ± 0.033 |
2.264 ± 0.014 | 5.029 ± 0.016 |
3.289 ± 0.023 | 10.285 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.424 ± 0.012 | 2.928 ± 0.019 |
4.938 ± 0.017 | 3.549 ± 0.018 |
6.631 ± 0.03 | 5.998 ± 0.024 |
5.6 ± 0.026 | 7.852 ± 0.017 |
1.328 ± 0.011 | 2.434 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
