
Bos taurus papillomavirus 11
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Xipapillomavirus; Xipapillomavirus 1
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
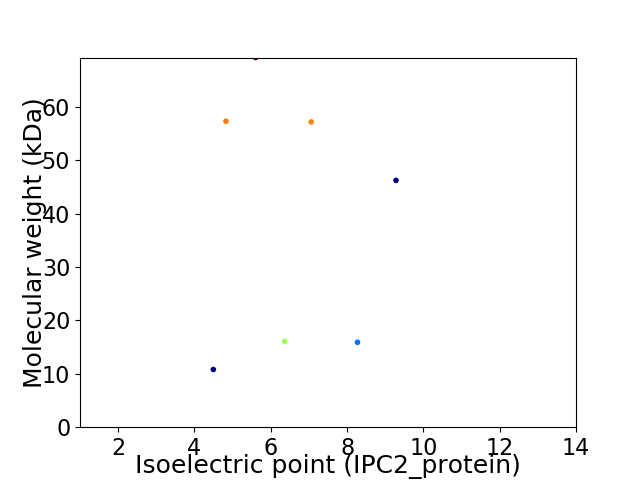
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E1CGB9|E1CGB9_BPV3 Replication protein E1 OS=Bos taurus papillomavirus 11 OX=714200 GN=E1 PE=3 SV=1
MM1 pKa = 7.64KK2 pKa = 10.31GPNVTLEE9 pKa = 4.6GIAADD14 pKa = 4.43LEE16 pKa = 4.64NAVSPINLNCEE27 pKa = 4.03EE28 pKa = 4.78EE29 pKa = 4.11IDD31 pKa = 3.97TEE33 pKa = 4.47EE34 pKa = 4.48VEE36 pKa = 4.49LPNPFEE42 pKa = 4.17ITATCYY48 pKa = 10.01VCEE51 pKa = 4.78KK52 pKa = 9.96ILRR55 pKa = 11.84LAVVTSNDD63 pKa = 4.67GIHH66 pKa = 6.35QLQQLLFEE74 pKa = 4.88NLSLLCSACSRR85 pKa = 11.84DD86 pKa = 3.58VFCNRR91 pKa = 11.84RR92 pKa = 11.84PVKK95 pKa = 10.6NGPP98 pKa = 3.32
MM1 pKa = 7.64KK2 pKa = 10.31GPNVTLEE9 pKa = 4.6GIAADD14 pKa = 4.43LEE16 pKa = 4.64NAVSPINLNCEE27 pKa = 4.03EE28 pKa = 4.78EE29 pKa = 4.11IDD31 pKa = 3.97TEE33 pKa = 4.47EE34 pKa = 4.48VEE36 pKa = 4.49LPNPFEE42 pKa = 4.17ITATCYY48 pKa = 10.01VCEE51 pKa = 4.78KK52 pKa = 9.96ILRR55 pKa = 11.84LAVVTSNDD63 pKa = 4.67GIHH66 pKa = 6.35QLQQLLFEE74 pKa = 4.88NLSLLCSACSRR85 pKa = 11.84DD86 pKa = 3.58VFCNRR91 pKa = 11.84RR92 pKa = 11.84PVKK95 pKa = 10.6NGPP98 pKa = 3.32
Molecular weight: 10.82 kDa
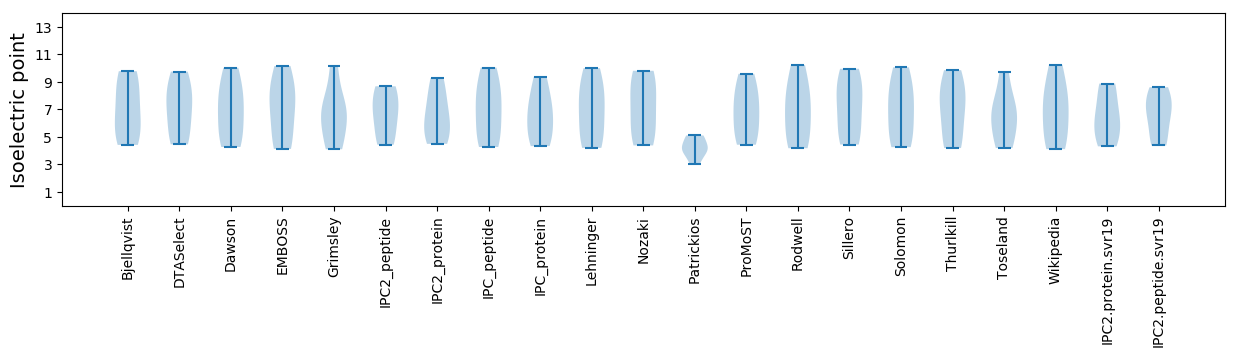
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E1CGC1|E1CGC1_BPV3 E4 protein OS=Bos taurus papillomavirus 11 OX=714200 GN=E4 PE=4 SV=1
MM1 pKa = 7.71ASLEE5 pKa = 4.03ARR7 pKa = 11.84FDD9 pKa = 3.74AVQEE13 pKa = 4.17SLLQIYY19 pKa = 9.7EE20 pKa = 4.37SEE22 pKa = 4.3SSSLEE27 pKa = 3.43LCYY30 pKa = 10.35QYY32 pKa = 10.46WQLIRR37 pKa = 11.84KK38 pKa = 8.28EE39 pKa = 3.74NALYY43 pKa = 8.26FTARR47 pKa = 11.84QQGKK51 pKa = 7.31TRR53 pKa = 11.84LGLYY57 pKa = 9.26AVPATRR63 pKa = 11.84VSEE66 pKa = 4.33QKK68 pKa = 11.0AKK70 pKa = 10.84DD71 pKa = 3.84AIKK74 pKa = 8.27MTLYY78 pKa = 10.92LEE80 pKa = 4.44SLKK83 pKa = 9.1QTQFATEE90 pKa = 3.87RR91 pKa = 11.84WTLMDD96 pKa = 3.68TSSEE100 pKa = 4.15TFMAPPEE107 pKa = 4.08NTFKK111 pKa = 10.99KK112 pKa = 9.89RR113 pKa = 11.84GQHH116 pKa = 3.64VTVVYY121 pKa = 10.8DD122 pKa = 3.62HH123 pKa = 7.53DD124 pKa = 5.12ANNSMLYY131 pKa = 8.97TLWTEE136 pKa = 4.46LYY138 pKa = 10.78YY139 pKa = 10.93LDD141 pKa = 5.72TSDD144 pKa = 3.58TWHH147 pKa = 6.62KK148 pKa = 6.72TTSSVDD154 pKa = 3.27YY155 pKa = 11.11DD156 pKa = 4.04GIFYY160 pKa = 10.23IDD162 pKa = 3.21IYY164 pKa = 8.95GTKK167 pKa = 9.03IYY169 pKa = 10.3YY170 pKa = 9.94INFQDD175 pKa = 5.02DD176 pKa = 3.2ASLYY180 pKa = 9.77SNSGQWEE187 pKa = 4.07VHH189 pKa = 5.73FEE191 pKa = 4.2NKK193 pKa = 9.46VLSPPVTSSLPVGTSGRR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84AQTGDD217 pKa = 2.87HH218 pKa = 6.23AQPGGRR224 pKa = 11.84KK225 pKa = 8.78PSQLAGDD232 pKa = 3.78SRR234 pKa = 11.84RR235 pKa = 11.84RR236 pKa = 11.84SQSQSSSRR244 pKa = 11.84SRR246 pKa = 11.84SRR248 pKa = 11.84SRR250 pKa = 11.84SPTKK254 pKa = 9.9EE255 pKa = 3.55PHH257 pKa = 5.21SGRR260 pKa = 11.84RR261 pKa = 11.84EE262 pKa = 3.79AGIPGQQQGRR272 pKa = 11.84PPGRR276 pKa = 11.84RR277 pKa = 11.84RR278 pKa = 11.84GEE280 pKa = 4.05SPEE283 pKa = 3.53QRR285 pKa = 11.84IFGGPSPPTPGQVGGRR301 pKa = 11.84HH302 pKa = 5.92RR303 pKa = 11.84SPSSKK308 pKa = 8.71PTSRR312 pKa = 11.84LARR315 pKa = 11.84LINEE319 pKa = 4.63AYY321 pKa = 10.02DD322 pKa = 3.8PPVLLLQGPANILKK336 pKa = 9.87CFRR339 pKa = 11.84RR340 pKa = 11.84RR341 pKa = 11.84ATQTHH346 pKa = 5.11GHH348 pKa = 6.74RR349 pKa = 11.84FLCMSTSWTWVCKK362 pKa = 9.63TSPLKK367 pKa = 10.61SGHH370 pKa = 6.3RR371 pKa = 11.84MLIAFTNCDD380 pKa = 3.01QRR382 pKa = 11.84KK383 pKa = 8.76GFLSSVRR390 pKa = 11.84LPKK393 pKa = 10.63GVTFVKK399 pKa = 10.8GSLTAYY405 pKa = 9.19SICYY409 pKa = 9.24
MM1 pKa = 7.71ASLEE5 pKa = 4.03ARR7 pKa = 11.84FDD9 pKa = 3.74AVQEE13 pKa = 4.17SLLQIYY19 pKa = 9.7EE20 pKa = 4.37SEE22 pKa = 4.3SSSLEE27 pKa = 3.43LCYY30 pKa = 10.35QYY32 pKa = 10.46WQLIRR37 pKa = 11.84KK38 pKa = 8.28EE39 pKa = 3.74NALYY43 pKa = 8.26FTARR47 pKa = 11.84QQGKK51 pKa = 7.31TRR53 pKa = 11.84LGLYY57 pKa = 9.26AVPATRR63 pKa = 11.84VSEE66 pKa = 4.33QKK68 pKa = 11.0AKK70 pKa = 10.84DD71 pKa = 3.84AIKK74 pKa = 8.27MTLYY78 pKa = 10.92LEE80 pKa = 4.44SLKK83 pKa = 9.1QTQFATEE90 pKa = 3.87RR91 pKa = 11.84WTLMDD96 pKa = 3.68TSSEE100 pKa = 4.15TFMAPPEE107 pKa = 4.08NTFKK111 pKa = 10.99KK112 pKa = 9.89RR113 pKa = 11.84GQHH116 pKa = 3.64VTVVYY121 pKa = 10.8DD122 pKa = 3.62HH123 pKa = 7.53DD124 pKa = 5.12ANNSMLYY131 pKa = 8.97TLWTEE136 pKa = 4.46LYY138 pKa = 10.78YY139 pKa = 10.93LDD141 pKa = 5.72TSDD144 pKa = 3.58TWHH147 pKa = 6.62KK148 pKa = 6.72TTSSVDD154 pKa = 3.27YY155 pKa = 11.11DD156 pKa = 4.04GIFYY160 pKa = 10.23IDD162 pKa = 3.21IYY164 pKa = 8.95GTKK167 pKa = 9.03IYY169 pKa = 10.3YY170 pKa = 9.94INFQDD175 pKa = 5.02DD176 pKa = 3.2ASLYY180 pKa = 9.77SNSGQWEE187 pKa = 4.07VHH189 pKa = 5.73FEE191 pKa = 4.2NKK193 pKa = 9.46VLSPPVTSSLPVGTSGRR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84AQTGDD217 pKa = 2.87HH218 pKa = 6.23AQPGGRR224 pKa = 11.84KK225 pKa = 8.78PSQLAGDD232 pKa = 3.78SRR234 pKa = 11.84RR235 pKa = 11.84RR236 pKa = 11.84SQSQSSSRR244 pKa = 11.84SRR246 pKa = 11.84SRR248 pKa = 11.84SRR250 pKa = 11.84SPTKK254 pKa = 9.9EE255 pKa = 3.55PHH257 pKa = 5.21SGRR260 pKa = 11.84RR261 pKa = 11.84EE262 pKa = 3.79AGIPGQQQGRR272 pKa = 11.84PPGRR276 pKa = 11.84RR277 pKa = 11.84RR278 pKa = 11.84GEE280 pKa = 4.05SPEE283 pKa = 3.53QRR285 pKa = 11.84IFGGPSPPTPGQVGGRR301 pKa = 11.84HH302 pKa = 5.92RR303 pKa = 11.84SPSSKK308 pKa = 8.71PTSRR312 pKa = 11.84LARR315 pKa = 11.84LINEE319 pKa = 4.63AYY321 pKa = 10.02DD322 pKa = 3.8PPVLLLQGPANILKK336 pKa = 9.87CFRR339 pKa = 11.84RR340 pKa = 11.84RR341 pKa = 11.84ATQTHH346 pKa = 5.11GHH348 pKa = 6.74RR349 pKa = 11.84FLCMSTSWTWVCKK362 pKa = 9.63TSPLKK367 pKa = 10.61SGHH370 pKa = 6.3RR371 pKa = 11.84MLIAFTNCDD380 pKa = 3.01QRR382 pKa = 11.84KK383 pKa = 8.76GFLSSVRR390 pKa = 11.84LPKK393 pKa = 10.63GVTFVKK399 pKa = 10.8GSLTAYY405 pKa = 9.19SICYY409 pKa = 9.24
Molecular weight: 46.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2424 |
98 |
606 |
346.3 |
38.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.817 ± 0.529 | 2.186 ± 0.531 |
5.899 ± 0.506 | 5.941 ± 0.622 |
4.703 ± 0.5 | 6.436 ± 0.715 |
2.269 ± 0.139 | 5.074 ± 0.585 |
4.827 ± 0.687 | 8.787 ± 0.974 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.939 ± 0.356 | 4.95 ± 0.755 |
6.477 ± 0.886 | 4.167 ± 0.458 |
6.559 ± 0.816 | 8.045 ± 0.789 |
5.899 ± 0.632 | 5.652 ± 0.636 |
1.526 ± 0.261 | 2.847 ± 0.377 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
