
Maricaulis maris (strain MCS10)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Maricaulales; Maricaulaceae; Maricaulis; Maricaulis maris
Average proteome isoelectric point is 5.85
Get precalculated fractions of proteins
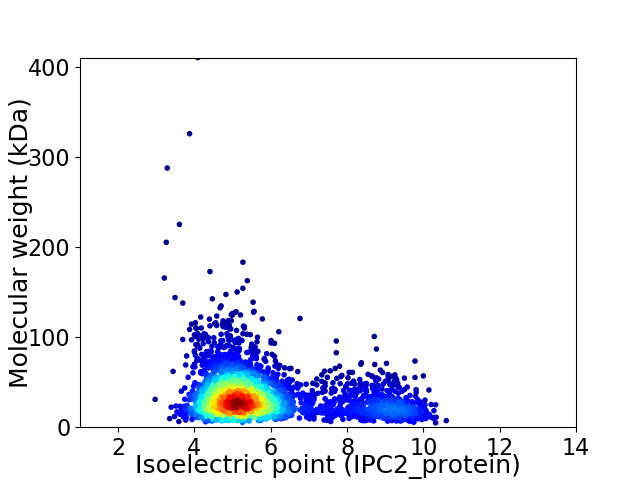
Virtual 2D-PAGE plot for 3060 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q0ANE1|SURE_MARMM 5'-nucleotidase SurE OS=Maricaulis maris (strain MCS10) OX=394221 GN=surE PE=3 SV=1
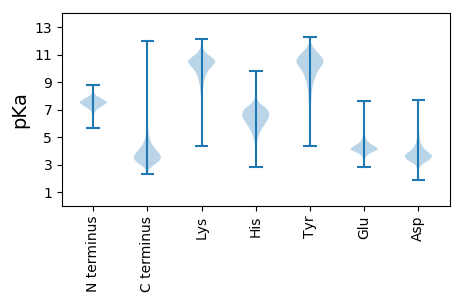
MM1 pKa = 7.27KK2 pKa = 9.73TFKK5 pKa = 10.82YY6 pKa = 10.82LMIGASALVLTACGNSDD23 pKa = 3.47TSRR26 pKa = 11.84DD27 pKa = 3.44LAEE30 pKa = 4.94ANVDD34 pKa = 3.66YY35 pKa = 10.83QATLMEE41 pKa = 4.31QLIDD45 pKa = 3.89AQPGDD50 pKa = 4.32VIEE53 pKa = 4.9IPAGVYY59 pKa = 10.55SFDD62 pKa = 4.23RR63 pKa = 11.84SLSLNIDD70 pKa = 3.36GVTIRR75 pKa = 11.84GAGMDD80 pKa = 3.59EE81 pKa = 4.73TILSFQDD88 pKa = 3.26QIAGAEE94 pKa = 4.14GLIVTANDD102 pKa = 3.28FTLEE106 pKa = 3.89NLAIEE111 pKa = 4.3DD112 pKa = 4.42TIGDD116 pKa = 3.93GLKK119 pKa = 10.96VNGGEE124 pKa = 4.08NIIIRR129 pKa = 11.84GVRR132 pKa = 11.84VEE134 pKa = 4.05WTNGYY139 pKa = 8.55ATEE142 pKa = 4.13NGAYY146 pKa = 10.01GIYY149 pKa = 9.89PVQTTNVLVEE159 pKa = 4.06DD160 pKa = 4.52TVAIGASDD168 pKa = 3.18AGIYY172 pKa = 9.7VGQSRR177 pKa = 11.84NVIVRR182 pKa = 11.84NSRR185 pKa = 11.84AEE187 pKa = 3.9YY188 pKa = 9.36NVAGIEE194 pKa = 3.97IEE196 pKa = 4.11NCIGADD202 pKa = 3.53VYY204 pKa = 11.79GNTATNNTGGILVFNMPNLPQPGYY228 pKa = 8.38HH229 pKa = 5.26TRR231 pKa = 11.84VYY233 pKa = 11.3DD234 pKa = 3.91NDD236 pKa = 2.89IFANNTEE243 pKa = 4.32NFGHH247 pKa = 7.3AGTPVASVPAGSGVVINSNDD267 pKa = 3.22QVEE270 pKa = 4.37IFNNRR275 pKa = 11.84ISDD278 pKa = 3.73NDD280 pKa = 3.72TANIIISSLHH290 pKa = 4.61STGYY294 pKa = 10.35SDD296 pKa = 5.63YY297 pKa = 11.41SVQTDD302 pKa = 4.0FDD304 pKa = 5.06PYY306 pKa = 10.66PEE308 pKa = 5.15SIWIHH313 pKa = 6.75DD314 pKa = 3.73NTYY317 pKa = 10.88SGGGTSPDD325 pKa = 3.77GLDD328 pKa = 3.27LTALKK333 pKa = 10.37IAMFGLNGALPDD345 pKa = 3.59VLFDD349 pKa = 5.13GFVDD353 pKa = 4.2DD354 pKa = 5.91AKK356 pKa = 11.12LVDD359 pKa = 4.12GALPDD364 pKa = 3.63ALRR367 pKa = 11.84ICVSDD372 pKa = 3.77PVDD375 pKa = 3.42VLNADD380 pKa = 3.85GPNGYY385 pKa = 10.34ASPSVDD391 pKa = 3.14TEE393 pKa = 4.23DD394 pKa = 4.41FRR396 pKa = 11.84CSLDD400 pKa = 3.37SLSAVEE406 pKa = 5.51LAFADD411 pKa = 3.72
MM1 pKa = 7.27KK2 pKa = 9.73TFKK5 pKa = 10.82YY6 pKa = 10.82LMIGASALVLTACGNSDD23 pKa = 3.47TSRR26 pKa = 11.84DD27 pKa = 3.44LAEE30 pKa = 4.94ANVDD34 pKa = 3.66YY35 pKa = 10.83QATLMEE41 pKa = 4.31QLIDD45 pKa = 3.89AQPGDD50 pKa = 4.32VIEE53 pKa = 4.9IPAGVYY59 pKa = 10.55SFDD62 pKa = 4.23RR63 pKa = 11.84SLSLNIDD70 pKa = 3.36GVTIRR75 pKa = 11.84GAGMDD80 pKa = 3.59EE81 pKa = 4.73TILSFQDD88 pKa = 3.26QIAGAEE94 pKa = 4.14GLIVTANDD102 pKa = 3.28FTLEE106 pKa = 3.89NLAIEE111 pKa = 4.3DD112 pKa = 4.42TIGDD116 pKa = 3.93GLKK119 pKa = 10.96VNGGEE124 pKa = 4.08NIIIRR129 pKa = 11.84GVRR132 pKa = 11.84VEE134 pKa = 4.05WTNGYY139 pKa = 8.55ATEE142 pKa = 4.13NGAYY146 pKa = 10.01GIYY149 pKa = 9.89PVQTTNVLVEE159 pKa = 4.06DD160 pKa = 4.52TVAIGASDD168 pKa = 3.18AGIYY172 pKa = 9.7VGQSRR177 pKa = 11.84NVIVRR182 pKa = 11.84NSRR185 pKa = 11.84AEE187 pKa = 3.9YY188 pKa = 9.36NVAGIEE194 pKa = 3.97IEE196 pKa = 4.11NCIGADD202 pKa = 3.53VYY204 pKa = 11.79GNTATNNTGGILVFNMPNLPQPGYY228 pKa = 8.38HH229 pKa = 5.26TRR231 pKa = 11.84VYY233 pKa = 11.3DD234 pKa = 3.91NDD236 pKa = 2.89IFANNTEE243 pKa = 4.32NFGHH247 pKa = 7.3AGTPVASVPAGSGVVINSNDD267 pKa = 3.22QVEE270 pKa = 4.37IFNNRR275 pKa = 11.84ISDD278 pKa = 3.73NDD280 pKa = 3.72TANIIISSLHH290 pKa = 4.61STGYY294 pKa = 10.35SDD296 pKa = 5.63YY297 pKa = 11.41SVQTDD302 pKa = 4.0FDD304 pKa = 5.06PYY306 pKa = 10.66PEE308 pKa = 5.15SIWIHH313 pKa = 6.75DD314 pKa = 3.73NTYY317 pKa = 10.88SGGGTSPDD325 pKa = 3.77GLDD328 pKa = 3.27LTALKK333 pKa = 10.37IAMFGLNGALPDD345 pKa = 3.59VLFDD349 pKa = 5.13GFVDD353 pKa = 4.2DD354 pKa = 5.91AKK356 pKa = 11.12LVDD359 pKa = 4.12GALPDD364 pKa = 3.63ALRR367 pKa = 11.84ICVSDD372 pKa = 3.77PVDD375 pKa = 3.42VLNADD380 pKa = 3.85GPNGYY385 pKa = 10.34ASPSVDD391 pKa = 3.14TEE393 pKa = 4.23DD394 pKa = 4.41FRR396 pKa = 11.84CSLDD400 pKa = 3.37SLSAVEE406 pKa = 5.51LAFADD411 pKa = 3.72
Molecular weight: 43.58 kDa
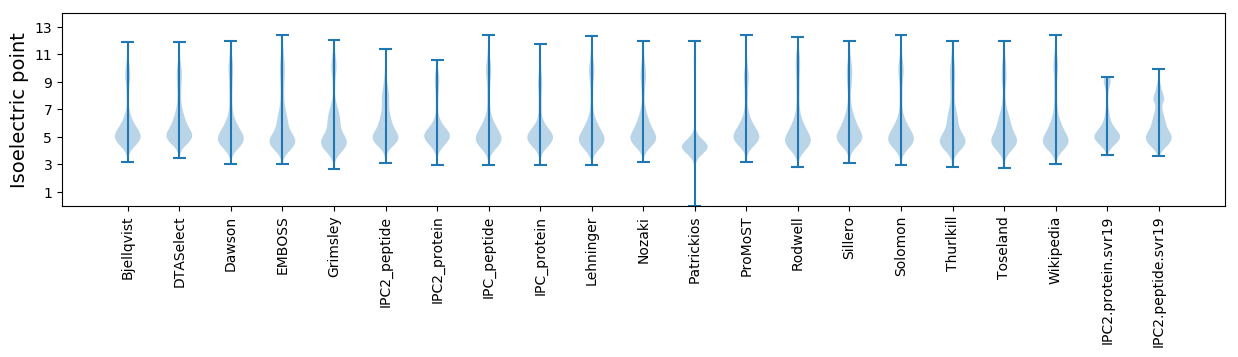
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q0ASZ6|Q0ASZ6_MARMM Proline racemase OS=Maricaulis maris (strain MCS10) OX=394221 GN=Mmar10_0298 PE=3 SV=1
MM1 pKa = 7.38TNVGHH6 pKa = 7.52KK7 pKa = 10.2KK8 pKa = 10.07RR9 pKa = 11.84IAPRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PRR17 pKa = 11.84HH18 pKa = 5.26PRR20 pKa = 11.84PMTRR24 pKa = 11.84PSPLQNRR31 pKa = 11.84VTPTGEE37 pKa = 3.69ICAHH41 pKa = 6.9AARR44 pKa = 11.84GTLMGNRR51 pKa = 11.84GILHH55 pKa = 7.33DD56 pKa = 4.45EE57 pKa = 4.08RR58 pKa = 11.84QQLGTARR65 pKa = 11.84WKK67 pKa = 8.72HH68 pKa = 4.65QAWVTCTLAFKK79 pKa = 10.62ARR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84DD84 pKa = 3.35LMKK87 pKa = 10.31PGNYY91 pKa = 8.88TEE93 pKa = 6.0LFFTDD98 pKa = 3.44EE99 pKa = 4.24AVALAAGHH107 pKa = 6.5RR108 pKa = 11.84PCGEE112 pKa = 3.92CRR114 pKa = 11.84RR115 pKa = 11.84EE116 pKa = 3.86DD117 pKa = 3.6FNRR120 pKa = 11.84FRR122 pKa = 11.84DD123 pKa = 4.06AFAKK127 pKa = 10.07AHH129 pKa = 7.12PDD131 pKa = 3.44LPHH134 pKa = 7.27RR135 pKa = 11.84APDD138 pKa = 3.54MDD140 pKa = 3.75RR141 pKa = 11.84LMHH144 pKa = 6.67KK145 pKa = 10.28ARR147 pKa = 11.84VVSRR151 pKa = 11.84SRR153 pKa = 11.84AQVTYY158 pKa = 10.64EE159 pKa = 4.33AMLSDD164 pKa = 3.84LPEE167 pKa = 4.12GVFFRR172 pKa = 11.84VAPAGGPLVRR182 pKa = 11.84WQGKK186 pKa = 7.23VLAWSFEE193 pKa = 4.29GYY195 pKa = 9.8SAGPDD200 pKa = 3.33ALPEE204 pKa = 4.01RR205 pKa = 11.84VEE207 pKa = 4.14VLTPEE212 pKa = 4.24PTVKK216 pKa = 10.59AIAAGYY222 pKa = 10.1VPVCIATGG230 pKa = 3.22
MM1 pKa = 7.38TNVGHH6 pKa = 7.52KK7 pKa = 10.2KK8 pKa = 10.07RR9 pKa = 11.84IAPRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PRR17 pKa = 11.84HH18 pKa = 5.26PRR20 pKa = 11.84PMTRR24 pKa = 11.84PSPLQNRR31 pKa = 11.84VTPTGEE37 pKa = 3.69ICAHH41 pKa = 6.9AARR44 pKa = 11.84GTLMGNRR51 pKa = 11.84GILHH55 pKa = 7.33DD56 pKa = 4.45EE57 pKa = 4.08RR58 pKa = 11.84QQLGTARR65 pKa = 11.84WKK67 pKa = 8.72HH68 pKa = 4.65QAWVTCTLAFKK79 pKa = 10.62ARR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84DD84 pKa = 3.35LMKK87 pKa = 10.31PGNYY91 pKa = 8.88TEE93 pKa = 6.0LFFTDD98 pKa = 3.44EE99 pKa = 4.24AVALAAGHH107 pKa = 6.5RR108 pKa = 11.84PCGEE112 pKa = 3.92CRR114 pKa = 11.84RR115 pKa = 11.84EE116 pKa = 3.86DD117 pKa = 3.6FNRR120 pKa = 11.84FRR122 pKa = 11.84DD123 pKa = 4.06AFAKK127 pKa = 10.07AHH129 pKa = 7.12PDD131 pKa = 3.44LPHH134 pKa = 7.27RR135 pKa = 11.84APDD138 pKa = 3.54MDD140 pKa = 3.75RR141 pKa = 11.84LMHH144 pKa = 6.67KK145 pKa = 10.28ARR147 pKa = 11.84VVSRR151 pKa = 11.84SRR153 pKa = 11.84AQVTYY158 pKa = 10.64EE159 pKa = 4.33AMLSDD164 pKa = 3.84LPEE167 pKa = 4.12GVFFRR172 pKa = 11.84VAPAGGPLVRR182 pKa = 11.84WQGKK186 pKa = 7.23VLAWSFEE193 pKa = 4.29GYY195 pKa = 9.8SAGPDD200 pKa = 3.33ALPEE204 pKa = 4.01RR205 pKa = 11.84VEE207 pKa = 4.14VLTPEE212 pKa = 4.24PTVKK216 pKa = 10.59AIAAGYY222 pKa = 10.1VPVCIATGG230 pKa = 3.22
Molecular weight: 25.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1006057 |
41 |
4368 |
328.8 |
35.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.701 ± 0.059 | 0.787 ± 0.015 |
6.623 ± 0.04 | 5.988 ± 0.047 |
3.734 ± 0.033 | 8.814 ± 0.097 |
1.957 ± 0.025 | 5.029 ± 0.03 |
2.49 ± 0.046 | 9.842 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.497 ± 0.025 | 2.595 ± 0.038 |
4.938 ± 0.035 | 3.061 ± 0.023 |
7.12 ± 0.055 | 5.65 ± 0.046 |
5.42 ± 0.046 | 7.177 ± 0.035 |
1.431 ± 0.022 | 2.147 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
