
Firmicutes bacterium CAG:822
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; environmental samples
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
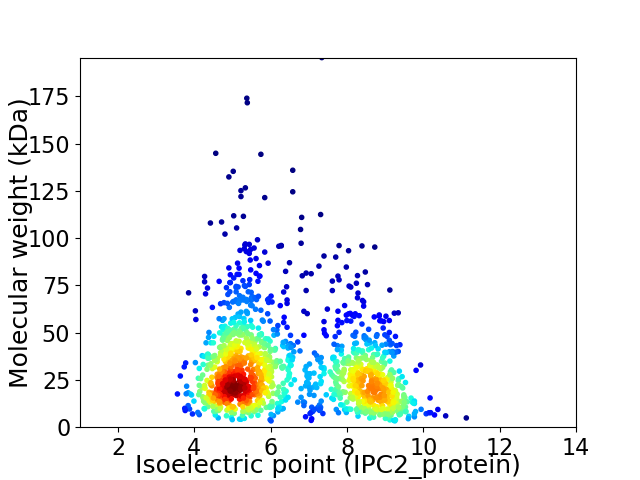
Virtual 2D-PAGE plot for 1377 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5IXR6|R5IXR6_9FIRM Rubredoxin-like domain-containing protein OS=Firmicutes bacterium CAG:822 OX=1263032 GN=BN793_01327 PE=4 SV=1
MM1 pKa = 7.42NEE3 pKa = 4.23NKK5 pKa = 10.14RR6 pKa = 11.84MLIFFISVVLVIAVILVIAFWPQSDD31 pKa = 3.77TTFACGVKK39 pKa = 10.14ADD41 pKa = 3.96SDD43 pKa = 4.12YY44 pKa = 11.87SRR46 pKa = 11.84LGSVNYY52 pKa = 9.17EE53 pKa = 3.92QYY55 pKa = 10.94SCLMEE60 pKa = 4.6QDD62 pKa = 4.18DD63 pKa = 5.03KK64 pKa = 11.53IALVVSDD71 pKa = 4.89GLNDD75 pKa = 4.49DD76 pKa = 4.39EE77 pKa = 5.7KK78 pKa = 11.5KK79 pKa = 10.83SLNDD83 pKa = 3.01VAEE86 pKa = 5.02RR87 pKa = 11.84ISKK90 pKa = 10.02SIYY93 pKa = 8.36YY94 pKa = 10.63LSDD97 pKa = 3.2EE98 pKa = 5.07VSNTDD103 pKa = 4.08LNNIKK108 pKa = 10.52DD109 pKa = 3.95DD110 pKa = 4.06LKK112 pKa = 10.21TDD114 pKa = 3.41EE115 pKa = 4.55VSYY118 pKa = 11.02DD119 pKa = 3.57NPSLVIIEE127 pKa = 4.24NGKK130 pKa = 8.19VTDD133 pKa = 5.48GIDD136 pKa = 3.41KK137 pKa = 11.0NLDD140 pKa = 3.47NSDD143 pKa = 3.9DD144 pKa = 4.12VYY146 pKa = 11.87DD147 pKa = 4.41FLQDD151 pKa = 2.94AGFAKK156 pKa = 9.96FACNATSDD164 pKa = 3.95SEE166 pKa = 4.47YY167 pKa = 10.9EE168 pKa = 3.9NLSRR172 pKa = 11.84LTYY175 pKa = 10.32DD176 pKa = 4.28QYY178 pKa = 11.78QCLYY182 pKa = 10.9DD183 pKa = 3.65SGEE186 pKa = 4.18PFVMILTQSTCSYY199 pKa = 9.89CAQFLPVINEE209 pKa = 3.99YY210 pKa = 10.82AGEE213 pKa = 4.2NNLPVYY219 pKa = 10.12FLEE222 pKa = 5.67IDD224 pKa = 3.56TMDD227 pKa = 3.99SDD229 pKa = 4.96DD230 pKa = 4.13LQSVFSSLSYY240 pKa = 10.85FSDD243 pKa = 3.26NSDD246 pKa = 2.57WGTPLTLGIKK256 pKa = 10.01DD257 pKa = 3.67KK258 pKa = 11.3EE259 pKa = 4.62VVTEE263 pKa = 4.01LSGYY267 pKa = 8.99TDD269 pKa = 3.55DD270 pKa = 4.19TSSIEE275 pKa = 4.04NLFTEE280 pKa = 5.36LGLKK284 pKa = 9.65
MM1 pKa = 7.42NEE3 pKa = 4.23NKK5 pKa = 10.14RR6 pKa = 11.84MLIFFISVVLVIAVILVIAFWPQSDD31 pKa = 3.77TTFACGVKK39 pKa = 10.14ADD41 pKa = 3.96SDD43 pKa = 4.12YY44 pKa = 11.87SRR46 pKa = 11.84LGSVNYY52 pKa = 9.17EE53 pKa = 3.92QYY55 pKa = 10.94SCLMEE60 pKa = 4.6QDD62 pKa = 4.18DD63 pKa = 5.03KK64 pKa = 11.53IALVVSDD71 pKa = 4.89GLNDD75 pKa = 4.49DD76 pKa = 4.39EE77 pKa = 5.7KK78 pKa = 11.5KK79 pKa = 10.83SLNDD83 pKa = 3.01VAEE86 pKa = 5.02RR87 pKa = 11.84ISKK90 pKa = 10.02SIYY93 pKa = 8.36YY94 pKa = 10.63LSDD97 pKa = 3.2EE98 pKa = 5.07VSNTDD103 pKa = 4.08LNNIKK108 pKa = 10.52DD109 pKa = 3.95DD110 pKa = 4.06LKK112 pKa = 10.21TDD114 pKa = 3.41EE115 pKa = 4.55VSYY118 pKa = 11.02DD119 pKa = 3.57NPSLVIIEE127 pKa = 4.24NGKK130 pKa = 8.19VTDD133 pKa = 5.48GIDD136 pKa = 3.41KK137 pKa = 11.0NLDD140 pKa = 3.47NSDD143 pKa = 3.9DD144 pKa = 4.12VYY146 pKa = 11.87DD147 pKa = 4.41FLQDD151 pKa = 2.94AGFAKK156 pKa = 9.96FACNATSDD164 pKa = 3.95SEE166 pKa = 4.47YY167 pKa = 10.9EE168 pKa = 3.9NLSRR172 pKa = 11.84LTYY175 pKa = 10.32DD176 pKa = 4.28QYY178 pKa = 11.78QCLYY182 pKa = 10.9DD183 pKa = 3.65SGEE186 pKa = 4.18PFVMILTQSTCSYY199 pKa = 9.89CAQFLPVINEE209 pKa = 3.99YY210 pKa = 10.82AGEE213 pKa = 4.2NNLPVYY219 pKa = 10.12FLEE222 pKa = 5.67IDD224 pKa = 3.56TMDD227 pKa = 3.99SDD229 pKa = 4.96DD230 pKa = 4.13LQSVFSSLSYY240 pKa = 10.85FSDD243 pKa = 3.26NSDD246 pKa = 2.57WGTPLTLGIKK256 pKa = 10.01DD257 pKa = 3.67KK258 pKa = 11.3EE259 pKa = 4.62VVTEE263 pKa = 4.01LSGYY267 pKa = 8.99TDD269 pKa = 3.55DD270 pKa = 4.19TSSIEE275 pKa = 4.04NLFTEE280 pKa = 5.36LGLKK284 pKa = 9.65
Molecular weight: 31.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5IVW1|R5IVW1_9FIRM Methyltransferase putative OS=Firmicutes bacterium CAG:822 OX=1263032 GN=BN793_00693 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPNTRR11 pKa = 11.84KK12 pKa = 9.8RR13 pKa = 11.84SKK15 pKa = 11.13KK16 pKa = 8.1MGFFARR22 pKa = 11.84MKK24 pKa = 10.39AGIVSRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 9.75KK34 pKa = 9.35GRR36 pKa = 11.84KK37 pKa = 8.41VLSHH41 pKa = 6.79
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPNTRR11 pKa = 11.84KK12 pKa = 9.8RR13 pKa = 11.84SKK15 pKa = 11.13KK16 pKa = 8.1MGFFARR22 pKa = 11.84MKK24 pKa = 10.39AGIVSRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 9.75KK34 pKa = 9.35GRR36 pKa = 11.84KK37 pKa = 8.41VLSHH41 pKa = 6.79
Molecular weight: 4.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
397830 |
29 |
1853 |
288.9 |
32.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.781 ± 0.067 | 0.994 ± 0.022 |
6.339 ± 0.056 | 7.14 ± 0.073 |
4.321 ± 0.064 | 5.629 ± 0.065 |
1.437 ± 0.031 | 9.427 ± 0.086 |
9.622 ± 0.092 | 9.144 ± 0.079 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.471 ± 0.026 | 7.026 ± 0.072 |
2.641 ± 0.036 | 2.3 ± 0.037 |
3.095 ± 0.045 | 6.122 ± 0.086 |
5.643 ± 0.103 | 6.19 ± 0.062 |
0.611 ± 0.02 | 5.066 ± 0.052 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
