
Bolahun virus variant 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Xinmoviridae; Anphevirus; Bolahun anphevirus; Bolahun virus
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
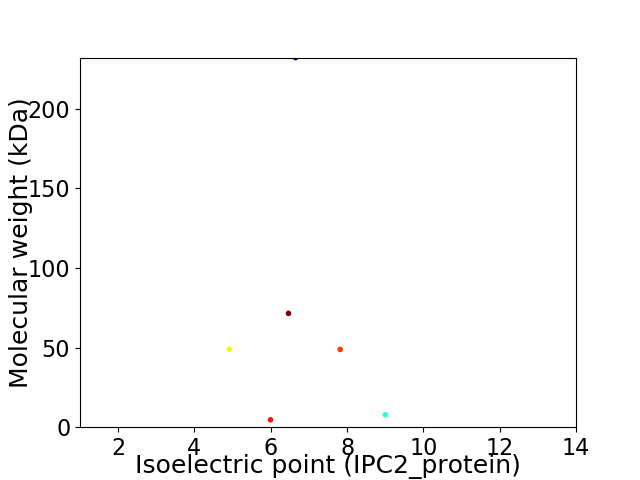
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1C9U5D8|A0A1C9U5D8_9MONO Putative small ZnF protein OS=Bolahun virus variant 2 OX=1903560 PE=4 SV=1
MM1 pKa = 8.14DD2 pKa = 4.07MALQQGNEE10 pKa = 4.15YY11 pKa = 10.33YY12 pKa = 10.18ISDD15 pKa = 4.68DD16 pKa = 4.62DD17 pKa = 5.03SDD19 pKa = 4.04GGPDD23 pKa = 3.49LFIPISQVDD32 pKa = 3.77TLRR35 pKa = 11.84EE36 pKa = 3.91MGFLPVSYY44 pKa = 10.19GHH46 pKa = 5.98YY47 pKa = 10.6SKK49 pKa = 10.43IRR51 pKa = 11.84KK52 pKa = 8.74NFLNEE57 pKa = 4.89FYY59 pKa = 9.98MAWAMAGTYY68 pKa = 10.07SAIVKK73 pKa = 7.43TNPSLSALDD82 pKa = 3.47ILSDD86 pKa = 3.63IEE88 pKa = 4.02IEE90 pKa = 4.35KK91 pKa = 10.82VGSCIMSDD99 pKa = 3.33EE100 pKa = 4.83GIIGCILLDD109 pKa = 4.22DD110 pKa = 4.51PRR112 pKa = 11.84AINTFKK118 pKa = 10.81DD119 pKa = 3.44LCSGRR124 pKa = 11.84GLFEE128 pKa = 3.85IVADD132 pKa = 4.24HH133 pKa = 5.85YY134 pKa = 11.74SEE136 pKa = 5.11VNMHH140 pKa = 6.26EE141 pKa = 4.43VSSMAPFCKK150 pKa = 10.22DD151 pKa = 3.03KK152 pKa = 11.43NDD154 pKa = 3.67AKK156 pKa = 11.05SWTNWTSTSRR166 pKa = 11.84SRR168 pKa = 11.84PNSVKK173 pKa = 10.47INFEE177 pKa = 3.92PLEE180 pKa = 4.58SINSFTVIMEE190 pKa = 4.1IRR192 pKa = 11.84AKK194 pKa = 10.63LDD196 pKa = 3.57LVSGASFDD204 pKa = 5.92LDD206 pKa = 3.23ILDD209 pKa = 4.13KK210 pKa = 11.45GKK212 pKa = 10.35VCSPVDD218 pKa = 3.43IRR220 pKa = 11.84PKK222 pKa = 10.29AKK224 pKa = 9.73VSEE227 pKa = 4.31VQEE230 pKa = 4.43VPTTSYY236 pKa = 10.87LPSAPSPSVTQPAVQNRR253 pKa = 11.84QITNNDD259 pKa = 3.16FQKK262 pKa = 10.82EE263 pKa = 4.09FVPTGHH269 pKa = 5.16VTEE272 pKa = 4.6VEE274 pKa = 4.35SFFNYY279 pKa = 9.25IMEE282 pKa = 4.3VRR284 pKa = 11.84ASPSTLGCGLMKK296 pKa = 10.38YY297 pKa = 10.01SPEE300 pKa = 4.53FIWSFALSNCVQGHH314 pKa = 5.41APSWARR320 pKa = 11.84NAPVLEE326 pKa = 4.97LISRR330 pKa = 11.84LNDD333 pKa = 3.27YY334 pKa = 10.16CIHH337 pKa = 6.36SIKK340 pKa = 10.77DD341 pKa = 2.92EE342 pKa = 4.1GMYY345 pKa = 10.13SRR347 pKa = 11.84EE348 pKa = 3.74MLGTKK353 pKa = 9.53TYY355 pKa = 10.79VDD357 pKa = 3.76SSTLSEE363 pKa = 4.08TMEE366 pKa = 4.41SLKK369 pKa = 10.77SSQKK373 pKa = 10.13RR374 pKa = 11.84EE375 pKa = 3.84SKK377 pKa = 10.75AILNVEE383 pKa = 4.2SMMKK387 pKa = 10.22EE388 pKa = 3.69LLAHH392 pKa = 6.54IKK394 pKa = 10.29SGEE397 pKa = 3.93RR398 pKa = 11.84SIPMAALPTVSAEE411 pKa = 4.05DD412 pKa = 3.51KK413 pKa = 10.74PLFEE417 pKa = 6.99AEE419 pKa = 4.33GQRR422 pKa = 11.84NPQVTKK428 pKa = 9.78EE429 pKa = 3.8PTGIVFIPPPPVV441 pKa = 2.91
MM1 pKa = 8.14DD2 pKa = 4.07MALQQGNEE10 pKa = 4.15YY11 pKa = 10.33YY12 pKa = 10.18ISDD15 pKa = 4.68DD16 pKa = 4.62DD17 pKa = 5.03SDD19 pKa = 4.04GGPDD23 pKa = 3.49LFIPISQVDD32 pKa = 3.77TLRR35 pKa = 11.84EE36 pKa = 3.91MGFLPVSYY44 pKa = 10.19GHH46 pKa = 5.98YY47 pKa = 10.6SKK49 pKa = 10.43IRR51 pKa = 11.84KK52 pKa = 8.74NFLNEE57 pKa = 4.89FYY59 pKa = 9.98MAWAMAGTYY68 pKa = 10.07SAIVKK73 pKa = 7.43TNPSLSALDD82 pKa = 3.47ILSDD86 pKa = 3.63IEE88 pKa = 4.02IEE90 pKa = 4.35KK91 pKa = 10.82VGSCIMSDD99 pKa = 3.33EE100 pKa = 4.83GIIGCILLDD109 pKa = 4.22DD110 pKa = 4.51PRR112 pKa = 11.84AINTFKK118 pKa = 10.81DD119 pKa = 3.44LCSGRR124 pKa = 11.84GLFEE128 pKa = 3.85IVADD132 pKa = 4.24HH133 pKa = 5.85YY134 pKa = 11.74SEE136 pKa = 5.11VNMHH140 pKa = 6.26EE141 pKa = 4.43VSSMAPFCKK150 pKa = 10.22DD151 pKa = 3.03KK152 pKa = 11.43NDD154 pKa = 3.67AKK156 pKa = 11.05SWTNWTSTSRR166 pKa = 11.84SRR168 pKa = 11.84PNSVKK173 pKa = 10.47INFEE177 pKa = 3.92PLEE180 pKa = 4.58SINSFTVIMEE190 pKa = 4.1IRR192 pKa = 11.84AKK194 pKa = 10.63LDD196 pKa = 3.57LVSGASFDD204 pKa = 5.92LDD206 pKa = 3.23ILDD209 pKa = 4.13KK210 pKa = 11.45GKK212 pKa = 10.35VCSPVDD218 pKa = 3.43IRR220 pKa = 11.84PKK222 pKa = 10.29AKK224 pKa = 9.73VSEE227 pKa = 4.31VQEE230 pKa = 4.43VPTTSYY236 pKa = 10.87LPSAPSPSVTQPAVQNRR253 pKa = 11.84QITNNDD259 pKa = 3.16FQKK262 pKa = 10.82EE263 pKa = 4.09FVPTGHH269 pKa = 5.16VTEE272 pKa = 4.6VEE274 pKa = 4.35SFFNYY279 pKa = 9.25IMEE282 pKa = 4.3VRR284 pKa = 11.84ASPSTLGCGLMKK296 pKa = 10.38YY297 pKa = 10.01SPEE300 pKa = 4.53FIWSFALSNCVQGHH314 pKa = 5.41APSWARR320 pKa = 11.84NAPVLEE326 pKa = 4.97LISRR330 pKa = 11.84LNDD333 pKa = 3.27YY334 pKa = 10.16CIHH337 pKa = 6.36SIKK340 pKa = 10.77DD341 pKa = 2.92EE342 pKa = 4.1GMYY345 pKa = 10.13SRR347 pKa = 11.84EE348 pKa = 3.74MLGTKK353 pKa = 9.53TYY355 pKa = 10.79VDD357 pKa = 3.76SSTLSEE363 pKa = 4.08TMEE366 pKa = 4.41SLKK369 pKa = 10.77SSQKK373 pKa = 10.13RR374 pKa = 11.84EE375 pKa = 3.84SKK377 pKa = 10.75AILNVEE383 pKa = 4.2SMMKK387 pKa = 10.22EE388 pKa = 3.69LLAHH392 pKa = 6.54IKK394 pKa = 10.29SGEE397 pKa = 3.93RR398 pKa = 11.84SIPMAALPTVSAEE411 pKa = 4.05DD412 pKa = 3.51KK413 pKa = 10.74PLFEE417 pKa = 6.99AEE419 pKa = 4.33GQRR422 pKa = 11.84NPQVTKK428 pKa = 9.78EE429 pKa = 3.8PTGIVFIPPPPVV441 pKa = 2.91
Molecular weight: 48.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1C9U5H2|A0A1C9U5H2_9MONO Putative glycoprotein OS=Bolahun virus variant 2 OX=1903560 PE=4 SV=1
MM1 pKa = 7.7AFYY4 pKa = 10.11FGPVIDD10 pKa = 4.08WQDD13 pKa = 3.25FLSFYY18 pKa = 9.83WLHH21 pKa = 6.44CFFVVIIIFILGLVVSLLRR40 pKa = 11.84GLTRR44 pKa = 11.84EE45 pKa = 3.92VRR47 pKa = 11.84EE48 pKa = 4.23PAWYY52 pKa = 8.97AQWVRR57 pKa = 11.84EE58 pKa = 4.24SKK60 pKa = 10.17SRR62 pKa = 11.84KK63 pKa = 8.49RR64 pKa = 3.52
MM1 pKa = 7.7AFYY4 pKa = 10.11FGPVIDD10 pKa = 4.08WQDD13 pKa = 3.25FLSFYY18 pKa = 9.83WLHH21 pKa = 6.44CFFVVIIIFILGLVVSLLRR40 pKa = 11.84GLTRR44 pKa = 11.84EE45 pKa = 3.92VRR47 pKa = 11.84EE48 pKa = 4.23PAWYY52 pKa = 8.97AQWVRR57 pKa = 11.84EE58 pKa = 4.24SKK60 pKa = 10.17SRR62 pKa = 11.84KK63 pKa = 8.49RR64 pKa = 3.52
Molecular weight: 7.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3647 |
40 |
2018 |
607.8 |
68.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.182 ± 0.68 | 2.468 ± 0.356 |
5.786 ± 0.377 | 6.307 ± 0.337 |
4.442 ± 0.275 | 5.84 ± 0.35 |
2.358 ± 0.447 | 6.828 ± 0.453 |
6.032 ± 0.247 | 9.158 ± 0.492 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.852 ± 0.282 | 4.552 ± 0.355 |
4.415 ± 0.636 | 2.906 ± 0.214 |
5.758 ± 0.784 | 8.199 ± 0.615 |
5.621 ± 0.716 | 6.252 ± 0.953 |
1.069 ± 0.185 | 3.976 ± 0.207 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
