
Enhygromyxa salina
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Myxococcales incertae sedis; Enhygromyxa
Average proteome isoelectric point is 5.82
Get precalculated fractions of proteins
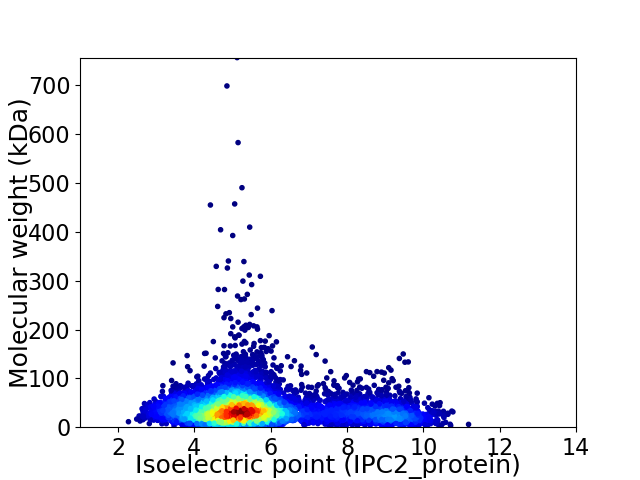
Virtual 2D-PAGE plot for 6943 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S9XLW3|A0A2S9XLW3_9DELT Uncharacterized protein OS=Enhygromyxa salina OX=215803 GN=ENSA5_42460 PE=4 SV=1
MM1 pKa = 7.24TNSLRR6 pKa = 11.84LLFGTCLILSPIACGDD22 pKa = 3.39SSATDD27 pKa = 3.63GEE29 pKa = 4.92GASEE33 pKa = 4.0TGYY36 pKa = 11.53GDD38 pKa = 3.51GDD40 pKa = 4.01GDD42 pKa = 4.31GDD44 pKa = 4.42PGDD47 pKa = 4.42GDD49 pKa = 4.49GDD51 pKa = 4.17PGDD54 pKa = 4.42GDD56 pKa = 4.49GDD58 pKa = 4.22PGDD61 pKa = 4.85GDD63 pKa = 3.96TGDD66 pKa = 4.53GDD68 pKa = 4.05TGDD71 pKa = 4.61GDD73 pKa = 4.05TGDD76 pKa = 4.61GDD78 pKa = 4.0TGDD81 pKa = 4.7GDD83 pKa = 4.2MDD85 pKa = 4.78CGPIGEE91 pKa = 5.24PPPEE95 pKa = 4.8APIVMVAPADD105 pKa = 3.89AGALPQMVAEE115 pKa = 4.88AEE117 pKa = 4.26PNTTFMFADD126 pKa = 4.18GTYY129 pKa = 10.0DD130 pKa = 3.78LSGGSQIQVRR140 pKa = 11.84TPGLRR145 pKa = 11.84FVGASNDD152 pKa = 3.34RR153 pKa = 11.84DD154 pKa = 4.02AVILDD159 pKa = 3.42ADD161 pKa = 3.98YY162 pKa = 11.31GIGEE166 pKa = 4.39IFLVTASDD174 pKa = 3.4TTIAHH179 pKa = 5.63MTLQRR184 pKa = 11.84ATWHH188 pKa = 6.88PIHH191 pKa = 6.13VTGGDD196 pKa = 3.41QANTEE201 pKa = 3.71NTMIYY206 pKa = 10.1DD207 pKa = 3.52VAVIDD212 pKa = 4.88PGEE215 pKa = 3.8QAIKK219 pKa = 10.45INASGAGYY227 pKa = 10.05FSDD230 pKa = 4.4YY231 pKa = 10.07GTVACSSVVMTDD243 pKa = 2.88AGRR246 pKa = 11.84AQVQNCYY253 pKa = 9.84TGGVDD258 pKa = 3.13AHH260 pKa = 6.16SARR263 pKa = 11.84GWVVRR268 pKa = 11.84DD269 pKa = 3.44NYY271 pKa = 10.91FEE273 pKa = 5.64GFWCDD278 pKa = 3.09VGLSEE283 pKa = 5.32HH284 pKa = 6.9AVHH287 pKa = 6.94FWITGRR293 pKa = 11.84DD294 pKa = 3.36TVVEE298 pKa = 4.19RR299 pKa = 11.84NTIVDD304 pKa = 3.87CARR307 pKa = 11.84GVGFGLGQNGNGKK320 pKa = 8.88QRR322 pKa = 11.84VYY324 pKa = 11.42DD325 pKa = 4.56DD326 pKa = 5.12DD327 pKa = 4.56PCPGADD333 pKa = 3.53YY334 pKa = 10.5LGHH337 pKa = 6.06VDD339 pKa = 5.09GVIRR343 pKa = 11.84DD344 pKa = 3.69NMIFAGRR351 pKa = 11.84PEE353 pKa = 4.46LFASQSGFDD362 pKa = 3.46SGVALEE368 pKa = 4.31QACGTVVAHH377 pKa = 5.58NTVASLQPPFVAMEE391 pKa = 3.84YY392 pKa = 10.31RR393 pKa = 11.84FPNTDD398 pKa = 2.64ATIVNNLTTHH408 pKa = 6.83GIVMRR413 pKa = 11.84DD414 pKa = 3.16GGQASLVGNLEE425 pKa = 4.18DD426 pKa = 3.71QGLEE430 pKa = 4.14HH431 pKa = 6.77FVDD434 pKa = 3.72AAGGDD439 pKa = 3.55LHH441 pKa = 8.33LVEE444 pKa = 5.9GSAAVDD450 pKa = 3.29AGDD453 pKa = 3.82PSGVDD458 pKa = 3.39WAPTDD463 pKa = 3.59YY464 pKa = 11.48DD465 pKa = 3.49GDD467 pKa = 4.1ARR469 pKa = 11.84DD470 pKa = 3.69GAPDD474 pKa = 3.38VGADD478 pKa = 3.78EE479 pKa = 5.22LPP481 pKa = 3.88
MM1 pKa = 7.24TNSLRR6 pKa = 11.84LLFGTCLILSPIACGDD22 pKa = 3.39SSATDD27 pKa = 3.63GEE29 pKa = 4.92GASEE33 pKa = 4.0TGYY36 pKa = 11.53GDD38 pKa = 3.51GDD40 pKa = 4.01GDD42 pKa = 4.31GDD44 pKa = 4.42PGDD47 pKa = 4.42GDD49 pKa = 4.49GDD51 pKa = 4.17PGDD54 pKa = 4.42GDD56 pKa = 4.49GDD58 pKa = 4.22PGDD61 pKa = 4.85GDD63 pKa = 3.96TGDD66 pKa = 4.53GDD68 pKa = 4.05TGDD71 pKa = 4.61GDD73 pKa = 4.05TGDD76 pKa = 4.61GDD78 pKa = 4.0TGDD81 pKa = 4.7GDD83 pKa = 4.2MDD85 pKa = 4.78CGPIGEE91 pKa = 5.24PPPEE95 pKa = 4.8APIVMVAPADD105 pKa = 3.89AGALPQMVAEE115 pKa = 4.88AEE117 pKa = 4.26PNTTFMFADD126 pKa = 4.18GTYY129 pKa = 10.0DD130 pKa = 3.78LSGGSQIQVRR140 pKa = 11.84TPGLRR145 pKa = 11.84FVGASNDD152 pKa = 3.34RR153 pKa = 11.84DD154 pKa = 4.02AVILDD159 pKa = 3.42ADD161 pKa = 3.98YY162 pKa = 11.31GIGEE166 pKa = 4.39IFLVTASDD174 pKa = 3.4TTIAHH179 pKa = 5.63MTLQRR184 pKa = 11.84ATWHH188 pKa = 6.88PIHH191 pKa = 6.13VTGGDD196 pKa = 3.41QANTEE201 pKa = 3.71NTMIYY206 pKa = 10.1DD207 pKa = 3.52VAVIDD212 pKa = 4.88PGEE215 pKa = 3.8QAIKK219 pKa = 10.45INASGAGYY227 pKa = 10.05FSDD230 pKa = 4.4YY231 pKa = 10.07GTVACSSVVMTDD243 pKa = 2.88AGRR246 pKa = 11.84AQVQNCYY253 pKa = 9.84TGGVDD258 pKa = 3.13AHH260 pKa = 6.16SARR263 pKa = 11.84GWVVRR268 pKa = 11.84DD269 pKa = 3.44NYY271 pKa = 10.91FEE273 pKa = 5.64GFWCDD278 pKa = 3.09VGLSEE283 pKa = 5.32HH284 pKa = 6.9AVHH287 pKa = 6.94FWITGRR293 pKa = 11.84DD294 pKa = 3.36TVVEE298 pKa = 4.19RR299 pKa = 11.84NTIVDD304 pKa = 3.87CARR307 pKa = 11.84GVGFGLGQNGNGKK320 pKa = 8.88QRR322 pKa = 11.84VYY324 pKa = 11.42DD325 pKa = 4.56DD326 pKa = 5.12DD327 pKa = 4.56PCPGADD333 pKa = 3.53YY334 pKa = 10.5LGHH337 pKa = 6.06VDD339 pKa = 5.09GVIRR343 pKa = 11.84DD344 pKa = 3.69NMIFAGRR351 pKa = 11.84PEE353 pKa = 4.46LFASQSGFDD362 pKa = 3.46SGVALEE368 pKa = 4.31QACGTVVAHH377 pKa = 5.58NTVASLQPPFVAMEE391 pKa = 3.84YY392 pKa = 10.31RR393 pKa = 11.84FPNTDD398 pKa = 2.64ATIVNNLTTHH408 pKa = 6.83GIVMRR413 pKa = 11.84DD414 pKa = 3.16GGQASLVGNLEE425 pKa = 4.18DD426 pKa = 3.71QGLEE430 pKa = 4.14HH431 pKa = 6.77FVDD434 pKa = 3.72AAGGDD439 pKa = 3.55LHH441 pKa = 8.33LVEE444 pKa = 5.9GSAAVDD450 pKa = 3.29AGDD453 pKa = 3.82PSGVDD458 pKa = 3.39WAPTDD463 pKa = 3.59YY464 pKa = 11.48DD465 pKa = 3.49GDD467 pKa = 4.1ARR469 pKa = 11.84DD470 pKa = 3.69GAPDD474 pKa = 3.38VGADD478 pKa = 3.78EE479 pKa = 5.22LPP481 pKa = 3.88
Molecular weight: 49.66 kDa
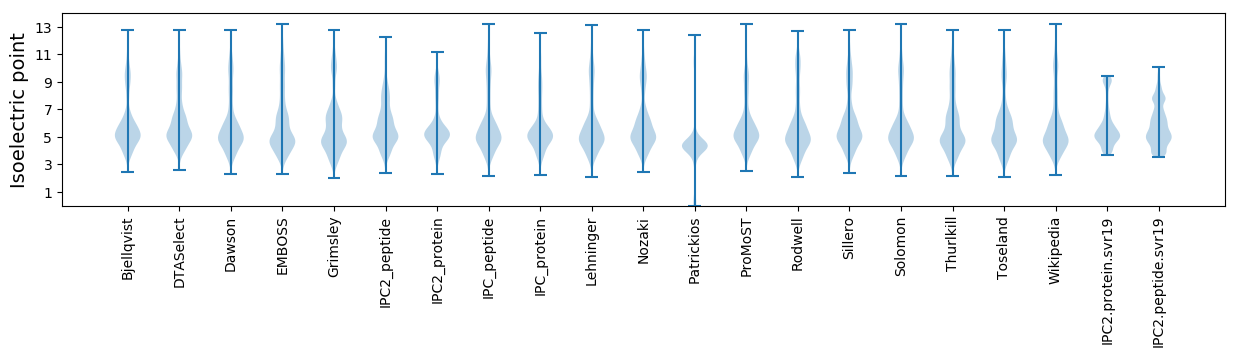
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S9XRF7|A0A2S9XRF7_9DELT 7 8-dihydroneopterin aldolase OS=Enhygromyxa salina OX=215803 GN=folB PE=3 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.96RR4 pKa = 11.84TYY6 pKa = 9.96QPHH9 pKa = 5.73NLSRR13 pKa = 11.84ARR15 pKa = 11.84THH17 pKa = 6.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MATRR26 pKa = 11.84NGRR29 pKa = 11.84KK30 pKa = 9.13IIAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.12RR42 pKa = 11.84LGTTTGGKK50 pKa = 9.53
MM1 pKa = 7.74SKK3 pKa = 8.96RR4 pKa = 11.84TYY6 pKa = 9.96QPHH9 pKa = 5.73NLSRR13 pKa = 11.84ARR15 pKa = 11.84THH17 pKa = 6.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MATRR26 pKa = 11.84NGRR29 pKa = 11.84KK30 pKa = 9.13IIAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.12RR42 pKa = 11.84LGTTTGGKK50 pKa = 9.53
Molecular weight: 5.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2649706 |
29 |
7134 |
381.6 |
41.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.235 ± 0.051 | 1.274 ± 0.016 |
6.693 ± 0.032 | 7.16 ± 0.035 |
3.053 ± 0.022 | 8.983 ± 0.041 |
2.165 ± 0.013 | 3.813 ± 0.019 |
2.315 ± 0.028 | 10.6 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.782 ± 0.012 | 2.003 ± 0.018 |
5.84 ± 0.024 | 2.936 ± 0.017 |
7.864 ± 0.043 | 5.479 ± 0.021 |
4.809 ± 0.021 | 7.488 ± 0.026 |
1.507 ± 0.012 | 2.0 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
