
[Clostridium] symbiosum WAL-14163
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Lachnoclostridium; [Clostridium] symbiosum
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
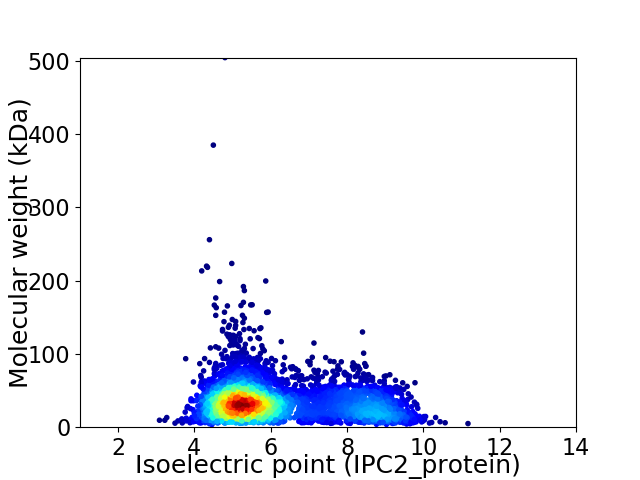
Virtual 2D-PAGE plot for 4668 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E7GNS1|E7GNS1_CLOSY Uncharacterized protein OS=[Clostridium] symbiosum WAL-14163 OX=742740 GN=HMPREF9474_02566 PE=4 SV=1
MM1 pKa = 7.43AASLTACSGDD11 pKa = 3.38SGTSGSGSTAAPAPAAEE28 pKa = 4.35SQGGEE33 pKa = 4.37GEE35 pKa = 4.68GVQAPSVDD43 pKa = 2.95IDD45 pKa = 3.47RR46 pKa = 11.84ATEE49 pKa = 3.96FVTIATGPTSGIYY62 pKa = 10.2YY63 pKa = 9.29PIGGAFATVLGNAGYY78 pKa = 7.86KK79 pKa = 9.67TSAQATGASVEE90 pKa = 4.22NINLITAGEE99 pKa = 4.18AEE101 pKa = 4.81LAIAMQDD108 pKa = 3.16SVMQAYY114 pKa = 9.75EE115 pKa = 4.03GFGAFEE121 pKa = 4.26TPNTEE126 pKa = 2.93LRR128 pKa = 11.84AAMRR132 pKa = 11.84LWPNYY137 pKa = 8.35VQLVTTANTGIKK149 pKa = 10.07SVEE152 pKa = 3.95DD153 pKa = 3.47LKK155 pKa = 11.38GKK157 pKa = 10.07RR158 pKa = 11.84VGVGAPNSGVEE169 pKa = 3.94LNARR173 pKa = 11.84MIYY176 pKa = 9.41EE177 pKa = 4.71AYY179 pKa = 10.32GMTYY183 pKa = 10.22EE184 pKa = 4.73DD185 pKa = 4.53SEE187 pKa = 4.43VDD189 pKa = 3.39YY190 pKa = 11.15LSYY193 pKa = 11.54GEE195 pKa = 6.07AIDD198 pKa = 4.03QMKK201 pKa = 10.68NGQCDD206 pKa = 3.21AAFVTSGLPNSTVSEE221 pKa = 4.16LAFSYY226 pKa = 11.55DD227 pKa = 3.58MVIVPIDD234 pKa = 3.4GDD236 pKa = 3.86GRR238 pKa = 11.84DD239 pKa = 3.75NLINKK244 pKa = 9.0YY245 pKa = 9.91PFFAASIIPADD256 pKa = 3.77TYY258 pKa = 11.75NNEE261 pKa = 3.86EE262 pKa = 4.08DD263 pKa = 3.83VEE265 pKa = 4.97SVFVYY270 pKa = 11.07NIMLVNKK277 pKa = 9.78DD278 pKa = 3.8LSNDD282 pKa = 3.25MVYY285 pKa = 11.64DD286 pKa = 3.6MMDD289 pKa = 4.37CIFNDD294 pKa = 2.96IGSIKK299 pKa = 10.25ASHH302 pKa = 5.88NTADD306 pKa = 3.49QNIDD310 pKa = 3.21VTFGVDD316 pKa = 3.25DD317 pKa = 4.1VKK319 pKa = 11.17IPLHH323 pKa = 6.94DD324 pKa = 4.75GAAKK328 pKa = 8.58WWQDD332 pKa = 2.8HH333 pKa = 7.02GYY335 pKa = 7.78DD336 pKa = 3.7TPQNN340 pKa = 3.62
MM1 pKa = 7.43AASLTACSGDD11 pKa = 3.38SGTSGSGSTAAPAPAAEE28 pKa = 4.35SQGGEE33 pKa = 4.37GEE35 pKa = 4.68GVQAPSVDD43 pKa = 2.95IDD45 pKa = 3.47RR46 pKa = 11.84ATEE49 pKa = 3.96FVTIATGPTSGIYY62 pKa = 10.2YY63 pKa = 9.29PIGGAFATVLGNAGYY78 pKa = 7.86KK79 pKa = 9.67TSAQATGASVEE90 pKa = 4.22NINLITAGEE99 pKa = 4.18AEE101 pKa = 4.81LAIAMQDD108 pKa = 3.16SVMQAYY114 pKa = 9.75EE115 pKa = 4.03GFGAFEE121 pKa = 4.26TPNTEE126 pKa = 2.93LRR128 pKa = 11.84AAMRR132 pKa = 11.84LWPNYY137 pKa = 8.35VQLVTTANTGIKK149 pKa = 10.07SVEE152 pKa = 3.95DD153 pKa = 3.47LKK155 pKa = 11.38GKK157 pKa = 10.07RR158 pKa = 11.84VGVGAPNSGVEE169 pKa = 3.94LNARR173 pKa = 11.84MIYY176 pKa = 9.41EE177 pKa = 4.71AYY179 pKa = 10.32GMTYY183 pKa = 10.22EE184 pKa = 4.73DD185 pKa = 4.53SEE187 pKa = 4.43VDD189 pKa = 3.39YY190 pKa = 11.15LSYY193 pKa = 11.54GEE195 pKa = 6.07AIDD198 pKa = 4.03QMKK201 pKa = 10.68NGQCDD206 pKa = 3.21AAFVTSGLPNSTVSEE221 pKa = 4.16LAFSYY226 pKa = 11.55DD227 pKa = 3.58MVIVPIDD234 pKa = 3.4GDD236 pKa = 3.86GRR238 pKa = 11.84DD239 pKa = 3.75NLINKK244 pKa = 9.0YY245 pKa = 9.91PFFAASIIPADD256 pKa = 3.77TYY258 pKa = 11.75NNEE261 pKa = 3.86EE262 pKa = 4.08DD263 pKa = 3.83VEE265 pKa = 4.97SVFVYY270 pKa = 11.07NIMLVNKK277 pKa = 9.78DD278 pKa = 3.8LSNDD282 pKa = 3.25MVYY285 pKa = 11.64DD286 pKa = 3.6MMDD289 pKa = 4.37CIFNDD294 pKa = 2.96IGSIKK299 pKa = 10.25ASHH302 pKa = 5.88NTADD306 pKa = 3.49QNIDD310 pKa = 3.21VTFGVDD316 pKa = 3.25DD317 pKa = 4.1VKK319 pKa = 11.17IPLHH323 pKa = 6.94DD324 pKa = 4.75GAAKK328 pKa = 8.58WWQDD332 pKa = 2.8HH333 pKa = 7.02GYY335 pKa = 7.78DD336 pKa = 3.7TPQNN340 pKa = 3.62
Molecular weight: 36.07 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E7GSD3|E7GSD3_CLOSY SpoJ protein OS=[Clostridium] symbiosum WAL-14163 OX=742740 GN=HMPREF9474_03828 PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.79KK9 pKa = 7.6RR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.44VHH16 pKa = 5.57GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSAGGRR28 pKa = 11.84KK29 pKa = 8.81VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.29GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.79KK9 pKa = 7.6RR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.44VHH16 pKa = 5.57GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSAGGRR28 pKa = 11.84KK29 pKa = 8.81VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.29GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 4.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1494359 |
31 |
4534 |
320.1 |
35.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.07 ± 0.041 | 1.511 ± 0.015 |
5.338 ± 0.028 | 7.555 ± 0.043 |
4.13 ± 0.029 | 7.756 ± 0.036 |
1.625 ± 0.013 | 7.098 ± 0.038 |
6.282 ± 0.029 | 9.142 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.168 ± 0.023 | 4.0 ± 0.025 |
3.585 ± 0.021 | 3.074 ± 0.023 |
4.893 ± 0.03 | 5.855 ± 0.027 |
5.347 ± 0.031 | 6.7 ± 0.03 |
0.941 ± 0.012 | 3.931 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
