
Dragonfly larvae associated circular virus-10
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.45
Get precalculated fractions of proteins
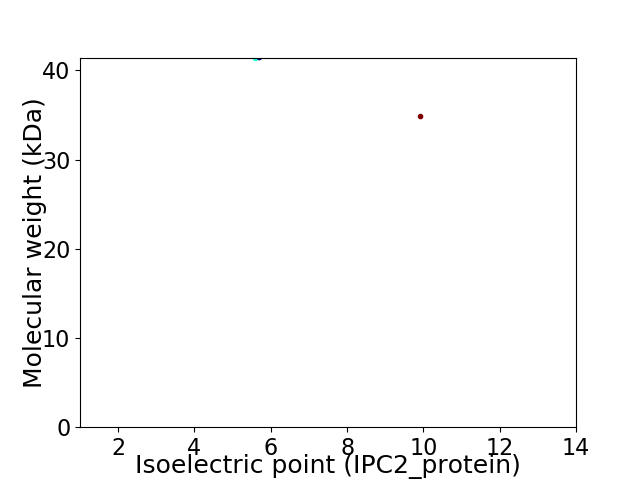
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5U2Q8|W5U2Q8_9VIRU Putative capsid protein OS=Dragonfly larvae associated circular virus-10 OX=1454022 PE=4 SV=1
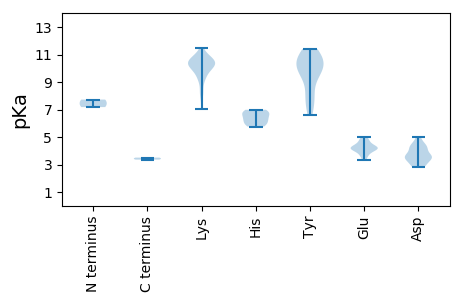
MM1 pKa = 7.72PKK3 pKa = 10.08VKK5 pKa = 10.17QGSQGEE11 pKa = 4.38RR12 pKa = 11.84WCFTLNNYY20 pKa = 7.23TEE22 pKa = 4.87KK23 pKa = 10.79DD24 pKa = 3.6VASITEE30 pKa = 4.15KK31 pKa = 10.4LTFSNCVFAKK41 pKa = 9.42VGKK44 pKa = 9.98EE45 pKa = 3.62IGDD48 pKa = 3.45SGTPHH53 pKa = 6.26LQGFIHH59 pKa = 6.22LRR61 pKa = 11.84KK62 pKa = 9.6RR63 pKa = 11.84LRR65 pKa = 11.84LACLKK70 pKa = 10.76KK71 pKa = 10.62LVGDD75 pKa = 4.08RR76 pKa = 11.84AHH78 pKa = 6.95CEE80 pKa = 3.91VARR83 pKa = 11.84GTDD86 pKa = 3.33KK87 pKa = 11.49DD88 pKa = 3.73NEE90 pKa = 4.68SYY92 pKa = 10.6CGKK95 pKa = 10.47DD96 pKa = 2.83SDD98 pKa = 4.25VVLTIGEE105 pKa = 4.24PTIGTTEE112 pKa = 3.8QGGGDD117 pKa = 4.2TIHH120 pKa = 6.98SIARR124 pKa = 11.84RR125 pKa = 11.84IALKK129 pKa = 10.4LSKK132 pKa = 10.22GTDD135 pKa = 2.86ITEE138 pKa = 4.21LQGEE142 pKa = 4.31EE143 pKa = 3.32WKK145 pKa = 10.68AYY147 pKa = 9.19CRR149 pKa = 11.84HH150 pKa = 5.88SKK152 pKa = 10.43VIQEE156 pKa = 4.14LSAALVKK163 pKa = 10.81NKK165 pKa = 10.2NIKK168 pKa = 9.91DD169 pKa = 3.5QAAQMSDD176 pKa = 2.82KK177 pKa = 10.67PLRR180 pKa = 11.84TWQKK184 pKa = 8.35EE185 pKa = 4.06LKK187 pKa = 10.29NAIQTVPDD195 pKa = 3.64DD196 pKa = 4.44RR197 pKa = 11.84KK198 pKa = 11.06VMWYY202 pKa = 9.56CDD204 pKa = 3.57SVGNTGKK211 pKa = 7.88TWFSKK216 pKa = 10.97YY217 pKa = 9.94LVALHH222 pKa = 5.77GAIRR226 pKa = 11.84FEE228 pKa = 4.3NGKK231 pKa = 9.72SADD234 pKa = 2.81IKK236 pKa = 10.1YY237 pKa = 9.96AYY239 pKa = 9.99NGEE242 pKa = 4.14RR243 pKa = 11.84VVVFDD248 pKa = 4.9LSRR251 pKa = 11.84SQVDD255 pKa = 3.12HH256 pKa = 6.69FNYY259 pKa = 10.21EE260 pKa = 4.09VIEE263 pKa = 4.41SIKK266 pKa = 10.44NGLMFSPKK274 pKa = 8.87YY275 pKa = 8.71TSCTKK280 pKa = 9.95MYY282 pKa = 8.29PIPHH286 pKa = 6.39VIVFANWMSDD296 pKa = 3.3EE297 pKa = 4.67SKK299 pKa = 10.9LSADD303 pKa = 2.99RR304 pKa = 11.84WKK306 pKa = 10.5IEE308 pKa = 4.03SLSDD312 pKa = 3.25VSKK315 pKa = 10.81IKK317 pKa = 10.55CEE319 pKa = 4.1KK320 pKa = 10.01EE321 pKa = 3.42DD322 pKa = 3.78DD323 pKa = 4.32VIVNDD328 pKa = 4.1VIVIEE333 pKa = 5.01DD334 pKa = 4.34DD335 pKa = 5.02DD336 pKa = 4.3VQQEE340 pKa = 4.27QIDD343 pKa = 4.44SYY345 pKa = 11.4FSNLFDD351 pKa = 3.43NWGPDD356 pKa = 3.47CVGDD360 pKa = 3.83LDD362 pKa = 4.73EE363 pKa = 4.52YY364 pKa = 11.15QQ365 pKa = 3.36
MM1 pKa = 7.72PKK3 pKa = 10.08VKK5 pKa = 10.17QGSQGEE11 pKa = 4.38RR12 pKa = 11.84WCFTLNNYY20 pKa = 7.23TEE22 pKa = 4.87KK23 pKa = 10.79DD24 pKa = 3.6VASITEE30 pKa = 4.15KK31 pKa = 10.4LTFSNCVFAKK41 pKa = 9.42VGKK44 pKa = 9.98EE45 pKa = 3.62IGDD48 pKa = 3.45SGTPHH53 pKa = 6.26LQGFIHH59 pKa = 6.22LRR61 pKa = 11.84KK62 pKa = 9.6RR63 pKa = 11.84LRR65 pKa = 11.84LACLKK70 pKa = 10.76KK71 pKa = 10.62LVGDD75 pKa = 4.08RR76 pKa = 11.84AHH78 pKa = 6.95CEE80 pKa = 3.91VARR83 pKa = 11.84GTDD86 pKa = 3.33KK87 pKa = 11.49DD88 pKa = 3.73NEE90 pKa = 4.68SYY92 pKa = 10.6CGKK95 pKa = 10.47DD96 pKa = 2.83SDD98 pKa = 4.25VVLTIGEE105 pKa = 4.24PTIGTTEE112 pKa = 3.8QGGGDD117 pKa = 4.2TIHH120 pKa = 6.98SIARR124 pKa = 11.84RR125 pKa = 11.84IALKK129 pKa = 10.4LSKK132 pKa = 10.22GTDD135 pKa = 2.86ITEE138 pKa = 4.21LQGEE142 pKa = 4.31EE143 pKa = 3.32WKK145 pKa = 10.68AYY147 pKa = 9.19CRR149 pKa = 11.84HH150 pKa = 5.88SKK152 pKa = 10.43VIQEE156 pKa = 4.14LSAALVKK163 pKa = 10.81NKK165 pKa = 10.2NIKK168 pKa = 9.91DD169 pKa = 3.5QAAQMSDD176 pKa = 2.82KK177 pKa = 10.67PLRR180 pKa = 11.84TWQKK184 pKa = 8.35EE185 pKa = 4.06LKK187 pKa = 10.29NAIQTVPDD195 pKa = 3.64DD196 pKa = 4.44RR197 pKa = 11.84KK198 pKa = 11.06VMWYY202 pKa = 9.56CDD204 pKa = 3.57SVGNTGKK211 pKa = 7.88TWFSKK216 pKa = 10.97YY217 pKa = 9.94LVALHH222 pKa = 5.77GAIRR226 pKa = 11.84FEE228 pKa = 4.3NGKK231 pKa = 9.72SADD234 pKa = 2.81IKK236 pKa = 10.1YY237 pKa = 9.96AYY239 pKa = 9.99NGEE242 pKa = 4.14RR243 pKa = 11.84VVVFDD248 pKa = 4.9LSRR251 pKa = 11.84SQVDD255 pKa = 3.12HH256 pKa = 6.69FNYY259 pKa = 10.21EE260 pKa = 4.09VIEE263 pKa = 4.41SIKK266 pKa = 10.44NGLMFSPKK274 pKa = 8.87YY275 pKa = 8.71TSCTKK280 pKa = 9.95MYY282 pKa = 8.29PIPHH286 pKa = 6.39VIVFANWMSDD296 pKa = 3.3EE297 pKa = 4.67SKK299 pKa = 10.9LSADD303 pKa = 2.99RR304 pKa = 11.84WKK306 pKa = 10.5IEE308 pKa = 4.03SLSDD312 pKa = 3.25VSKK315 pKa = 10.81IKK317 pKa = 10.55CEE319 pKa = 4.1KK320 pKa = 10.01EE321 pKa = 3.42DD322 pKa = 3.78DD323 pKa = 4.32VIVNDD328 pKa = 4.1VIVIEE333 pKa = 5.01DD334 pKa = 4.34DD335 pKa = 5.02DD336 pKa = 4.3VQQEE340 pKa = 4.27QIDD343 pKa = 4.44SYY345 pKa = 11.4FSNLFDD351 pKa = 3.43NWGPDD356 pKa = 3.47CVGDD360 pKa = 3.83LDD362 pKa = 4.73EE363 pKa = 4.52YY364 pKa = 11.15QQ365 pKa = 3.36
Molecular weight: 41.34 kDa
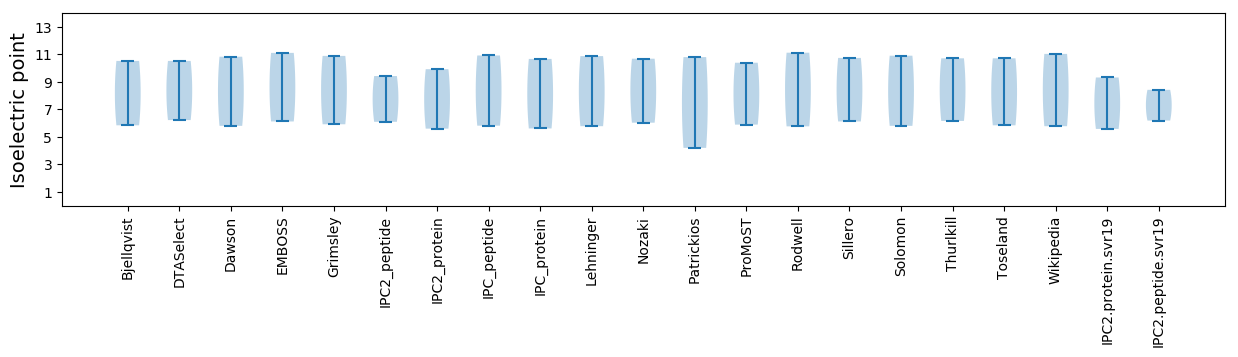
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5U2Q8|W5U2Q8_9VIRU Putative capsid protein OS=Dragonfly larvae associated circular virus-10 OX=1454022 PE=4 SV=1
MM1 pKa = 7.18IKK3 pKa = 10.0AYY5 pKa = 10.0RR6 pKa = 11.84VGRR9 pKa = 11.84EE10 pKa = 4.02RR11 pKa = 11.84IATTQLSAVTMAPYY25 pKa = 10.67RR26 pKa = 11.84NLTRR30 pKa = 11.84GRR32 pKa = 11.84YY33 pKa = 6.6SAKK36 pKa = 9.3RR37 pKa = 11.84RR38 pKa = 11.84YY39 pKa = 7.24RR40 pKa = 11.84TRR42 pKa = 11.84RR43 pKa = 11.84VTRR46 pKa = 11.84KK47 pKa = 9.09RR48 pKa = 11.84YY49 pKa = 9.49ARR51 pKa = 11.84TRR53 pKa = 11.84FSKK56 pKa = 10.52RR57 pKa = 11.84RR58 pKa = 11.84FGHH61 pKa = 6.65RR62 pKa = 11.84NRR64 pKa = 11.84SWAMRR69 pKa = 11.84KK70 pKa = 9.69RR71 pKa = 11.84ISNRR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84SGTKK81 pKa = 8.36VLKK84 pKa = 10.3VVHH87 pKa = 6.79RR88 pKa = 11.84WTIDD92 pKa = 2.89KK93 pKa = 10.93SFEE96 pKa = 3.74NGIYY100 pKa = 9.79TDD102 pKa = 4.25CLSMNASDD110 pKa = 4.55VNNLPGWAQLTVLYY124 pKa = 10.55DD125 pKa = 3.07EE126 pKa = 4.9WKK128 pKa = 10.1PGKK131 pKa = 10.19KK132 pKa = 9.77YY133 pKa = 9.87MKK135 pKa = 10.34SFVPFDD141 pKa = 3.32KK142 pKa = 11.4YY143 pKa = 11.25EE144 pKa = 4.29NEE146 pKa = 4.13VSDD149 pKa = 3.79NAEE152 pKa = 4.33LIVRR156 pKa = 11.84WSAYY160 pKa = 10.6DD161 pKa = 3.49PDD163 pKa = 3.99AKK165 pKa = 10.61GRR167 pKa = 11.84KK168 pKa = 7.07FTAARR173 pKa = 11.84AVSDD177 pKa = 3.73MEE179 pKa = 4.78KK180 pKa = 9.23MQNSKK185 pKa = 9.5WKK187 pKa = 10.19LIKK190 pKa = 10.29KK191 pKa = 8.09NQVVSTSYY199 pKa = 11.12QPMFPTRR206 pKa = 11.84TGVNLTGVKK215 pKa = 10.4GVDD218 pKa = 3.6NPWRR222 pKa = 11.84DD223 pKa = 3.53CVASVTSEE231 pKa = 3.68QCQNGVQQVWKK242 pKa = 10.48VNTPTTLVTRR252 pKa = 11.84HH253 pKa = 5.95YY254 pKa = 7.86FTQVYY259 pKa = 7.88YY260 pKa = 10.9FRR262 pKa = 11.84GLRR265 pKa = 11.84IGTQYY270 pKa = 10.39GAQYY274 pKa = 10.81EE275 pKa = 4.55PVTPAFPEE283 pKa = 4.17VVPPLKK289 pKa = 10.57KK290 pKa = 9.22VDD292 pKa = 3.48RR293 pKa = 11.84MRR295 pKa = 11.84TEE297 pKa = 3.5
MM1 pKa = 7.18IKK3 pKa = 10.0AYY5 pKa = 10.0RR6 pKa = 11.84VGRR9 pKa = 11.84EE10 pKa = 4.02RR11 pKa = 11.84IATTQLSAVTMAPYY25 pKa = 10.67RR26 pKa = 11.84NLTRR30 pKa = 11.84GRR32 pKa = 11.84YY33 pKa = 6.6SAKK36 pKa = 9.3RR37 pKa = 11.84RR38 pKa = 11.84YY39 pKa = 7.24RR40 pKa = 11.84TRR42 pKa = 11.84RR43 pKa = 11.84VTRR46 pKa = 11.84KK47 pKa = 9.09RR48 pKa = 11.84YY49 pKa = 9.49ARR51 pKa = 11.84TRR53 pKa = 11.84FSKK56 pKa = 10.52RR57 pKa = 11.84RR58 pKa = 11.84FGHH61 pKa = 6.65RR62 pKa = 11.84NRR64 pKa = 11.84SWAMRR69 pKa = 11.84KK70 pKa = 9.69RR71 pKa = 11.84ISNRR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84SGTKK81 pKa = 8.36VLKK84 pKa = 10.3VVHH87 pKa = 6.79RR88 pKa = 11.84WTIDD92 pKa = 2.89KK93 pKa = 10.93SFEE96 pKa = 3.74NGIYY100 pKa = 9.79TDD102 pKa = 4.25CLSMNASDD110 pKa = 4.55VNNLPGWAQLTVLYY124 pKa = 10.55DD125 pKa = 3.07EE126 pKa = 4.9WKK128 pKa = 10.1PGKK131 pKa = 10.19KK132 pKa = 9.77YY133 pKa = 9.87MKK135 pKa = 10.34SFVPFDD141 pKa = 3.32KK142 pKa = 11.4YY143 pKa = 11.25EE144 pKa = 4.29NEE146 pKa = 4.13VSDD149 pKa = 3.79NAEE152 pKa = 4.33LIVRR156 pKa = 11.84WSAYY160 pKa = 10.6DD161 pKa = 3.49PDD163 pKa = 3.99AKK165 pKa = 10.61GRR167 pKa = 11.84KK168 pKa = 7.07FTAARR173 pKa = 11.84AVSDD177 pKa = 3.73MEE179 pKa = 4.78KK180 pKa = 9.23MQNSKK185 pKa = 9.5WKK187 pKa = 10.19LIKK190 pKa = 10.29KK191 pKa = 8.09NQVVSTSYY199 pKa = 11.12QPMFPTRR206 pKa = 11.84TGVNLTGVKK215 pKa = 10.4GVDD218 pKa = 3.6NPWRR222 pKa = 11.84DD223 pKa = 3.53CVASVTSEE231 pKa = 3.68QCQNGVQQVWKK242 pKa = 10.48VNTPTTLVTRR252 pKa = 11.84HH253 pKa = 5.95YY254 pKa = 7.86FTQVYY259 pKa = 7.88YY260 pKa = 10.9FRR262 pKa = 11.84GLRR265 pKa = 11.84IGTQYY270 pKa = 10.39GAQYY274 pKa = 10.81EE275 pKa = 4.55PVTPAFPEE283 pKa = 4.17VVPPLKK289 pKa = 10.57KK290 pKa = 9.22VDD292 pKa = 3.48RR293 pKa = 11.84MRR295 pKa = 11.84TEE297 pKa = 3.5
Molecular weight: 34.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
662 |
297 |
365 |
331.0 |
38.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.589 ± 0.315 | 1.964 ± 0.638 |
6.495 ± 1.642 | 5.287 ± 1.059 |
3.323 ± 0.029 | 5.891 ± 0.562 |
1.662 ± 0.436 | 4.834 ± 1.431 |
8.761 ± 0.455 | 5.589 ± 0.811 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.266 ± 0.511 | 4.683 ± 0.246 |
3.474 ± 0.829 | 4.079 ± 0.026 |
7.553 ± 2.83 | 6.798 ± 0.493 |
6.647 ± 1.184 | 8.459 ± 0.648 |
2.417 ± 0.185 | 4.23 ± 0.774 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
