
Synechococcus sp. RS9916
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Synechococcaceae; Synechococcus; unclassified Synechococcus
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
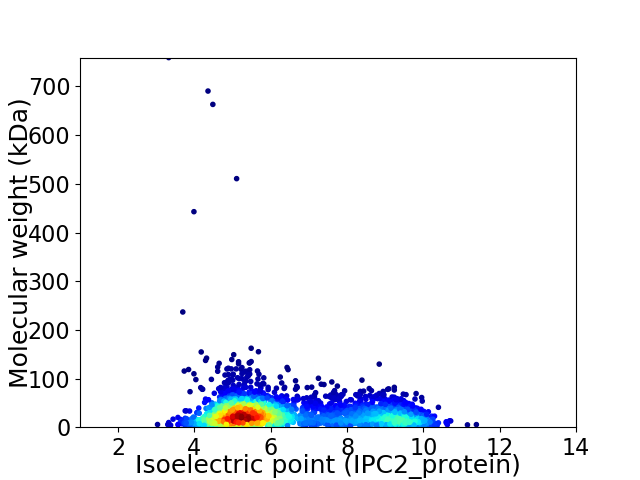
Virtual 2D-PAGE plot for 2955 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
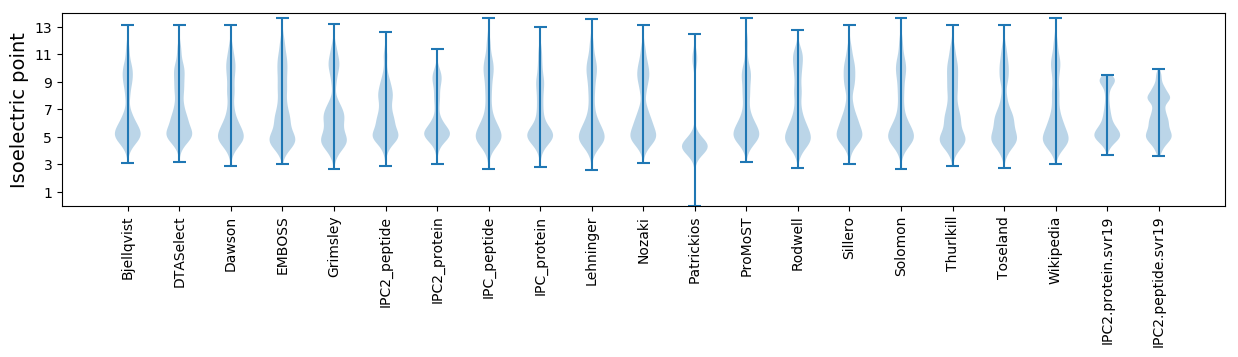
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q05SH5|Q05SH5_9SYNE Uncharacterized protein OS=Synechococcus sp. RS9916 OX=221359 GN=RS9916_25629 PE=4 SV=1
MM1 pKa = 7.79IPIKK5 pKa = 10.56GWGVDD10 pKa = 3.49HH11 pKa = 7.69GDD13 pKa = 3.84GEE15 pKa = 5.17CVGGRR20 pKa = 11.84PPQHH24 pKa = 6.64INPNNTMPFTFLNFNQKK41 pKa = 10.11LFFGDD46 pKa = 3.82SQDD49 pKa = 4.4DD50 pKa = 3.73AVLSLGSSKK59 pKa = 10.82VINLGGGNNTIYY71 pKa = 11.04SFGTNNTATSGSGDD85 pKa = 3.38DD86 pKa = 3.49RR87 pKa = 11.84FYY89 pKa = 10.83MIGGDD94 pKa = 3.47QTIVSTGGDD103 pKa = 3.22NTAITSSGDD112 pKa = 3.23DD113 pKa = 3.47RR114 pKa = 11.84IIFSNQIGGNDD125 pKa = 3.61YY126 pKa = 10.51VRR128 pKa = 11.84SGSGEE133 pKa = 3.94DD134 pKa = 3.42VIYY137 pKa = 10.5TSGGDD142 pKa = 3.81DD143 pKa = 3.69EE144 pKa = 4.83VHH146 pKa = 6.74SGADD150 pKa = 3.22NDD152 pKa = 3.84QVYY155 pKa = 10.45LGSGNDD161 pKa = 3.4TAYY164 pKa = 10.76LGSGDD169 pKa = 3.86DD170 pKa = 3.31VAYY173 pKa = 10.51AGTGTDD179 pKa = 5.09LIDD182 pKa = 4.93GEE184 pKa = 5.14DD185 pKa = 3.93GNDD188 pKa = 3.9TISGLHH194 pKa = 5.75SKK196 pKa = 10.55GNKK199 pKa = 7.7TFFGRR204 pKa = 11.84AGDD207 pKa = 3.8DD208 pKa = 3.47TLTGGVHH215 pKa = 6.83NDD217 pKa = 3.59YY218 pKa = 11.31LDD220 pKa = 4.01GGADD224 pKa = 3.53ADD226 pKa = 3.87VLTGNEE232 pKa = 4.15GFNIYY237 pKa = 9.34WGGDD241 pKa = 3.34DD242 pKa = 4.94ADD244 pKa = 3.61VFRR247 pKa = 11.84IGRR250 pKa = 11.84GVDD253 pKa = 4.02LIEE256 pKa = 5.19DD257 pKa = 4.06FNLAEE262 pKa = 4.82GDD264 pKa = 3.82TIEE267 pKa = 4.28MVEE270 pKa = 4.69PIASICASVSAHH282 pKa = 4.32VVNYY286 pKa = 10.64SGTLVNGQSISGSIVCEE303 pKa = 4.19TEE305 pKa = 3.39NQARR309 pKa = 11.84DD310 pKa = 4.18FYY312 pKa = 10.59FNWMPQASS320 pKa = 3.6
MM1 pKa = 7.79IPIKK5 pKa = 10.56GWGVDD10 pKa = 3.49HH11 pKa = 7.69GDD13 pKa = 3.84GEE15 pKa = 5.17CVGGRR20 pKa = 11.84PPQHH24 pKa = 6.64INPNNTMPFTFLNFNQKK41 pKa = 10.11LFFGDD46 pKa = 3.82SQDD49 pKa = 4.4DD50 pKa = 3.73AVLSLGSSKK59 pKa = 10.82VINLGGGNNTIYY71 pKa = 11.04SFGTNNTATSGSGDD85 pKa = 3.38DD86 pKa = 3.49RR87 pKa = 11.84FYY89 pKa = 10.83MIGGDD94 pKa = 3.47QTIVSTGGDD103 pKa = 3.22NTAITSSGDD112 pKa = 3.23DD113 pKa = 3.47RR114 pKa = 11.84IIFSNQIGGNDD125 pKa = 3.61YY126 pKa = 10.51VRR128 pKa = 11.84SGSGEE133 pKa = 3.94DD134 pKa = 3.42VIYY137 pKa = 10.5TSGGDD142 pKa = 3.81DD143 pKa = 3.69EE144 pKa = 4.83VHH146 pKa = 6.74SGADD150 pKa = 3.22NDD152 pKa = 3.84QVYY155 pKa = 10.45LGSGNDD161 pKa = 3.4TAYY164 pKa = 10.76LGSGDD169 pKa = 3.86DD170 pKa = 3.31VAYY173 pKa = 10.51AGTGTDD179 pKa = 5.09LIDD182 pKa = 4.93GEE184 pKa = 5.14DD185 pKa = 3.93GNDD188 pKa = 3.9TISGLHH194 pKa = 5.75SKK196 pKa = 10.55GNKK199 pKa = 7.7TFFGRR204 pKa = 11.84AGDD207 pKa = 3.8DD208 pKa = 3.47TLTGGVHH215 pKa = 6.83NDD217 pKa = 3.59YY218 pKa = 11.31LDD220 pKa = 4.01GGADD224 pKa = 3.53ADD226 pKa = 3.87VLTGNEE232 pKa = 4.15GFNIYY237 pKa = 9.34WGGDD241 pKa = 3.34DD242 pKa = 4.94ADD244 pKa = 3.61VFRR247 pKa = 11.84IGRR250 pKa = 11.84GVDD253 pKa = 4.02LIEE256 pKa = 5.19DD257 pKa = 4.06FNLAEE262 pKa = 4.82GDD264 pKa = 3.82TIEE267 pKa = 4.28MVEE270 pKa = 4.69PIASICASVSAHH282 pKa = 4.32VVNYY286 pKa = 10.64SGTLVNGQSISGSIVCEE303 pKa = 4.19TEE305 pKa = 3.39NQARR309 pKa = 11.84DD310 pKa = 4.18FYY312 pKa = 10.59FNWMPQASS320 pKa = 3.6
Molecular weight: 33.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q05WM3|Q05WM3_9SYNE Uncharacterized protein OS=Synechococcus sp. RS9916 OX=221359 GN=RS9916_33607 PE=4 SV=1
MM1 pKa = 7.55TKK3 pKa = 9.01RR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 9.07RR13 pKa = 11.84KK14 pKa = 8.17RR15 pKa = 11.84VSGFRR20 pKa = 11.84VRR22 pKa = 11.84MRR24 pKa = 11.84SHH26 pKa = 6.21TGRR29 pKa = 11.84RR30 pKa = 11.84VIRR33 pKa = 11.84TRR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.74RR38 pKa = 11.84GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.99
MM1 pKa = 7.55TKK3 pKa = 9.01RR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 9.07RR13 pKa = 11.84KK14 pKa = 8.17RR15 pKa = 11.84VSGFRR20 pKa = 11.84VRR22 pKa = 11.84MRR24 pKa = 11.84SHH26 pKa = 6.21TGRR29 pKa = 11.84RR30 pKa = 11.84VIRR33 pKa = 11.84TRR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.74RR38 pKa = 11.84GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.99
Molecular weight: 5.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
801681 |
20 |
7750 |
271.3 |
29.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.756 ± 0.063 | 1.185 ± 0.024 |
5.363 ± 0.051 | 5.458 ± 0.062 |
2.957 ± 0.04 | 8.83 ± 0.19 |
2.057 ± 0.039 | 4.177 ± 0.048 |
2.712 ± 0.048 | 12.063 ± 0.098 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.036 ± 0.03 | 2.913 ± 0.076 |
5.371 ± 0.072 | 4.868 ± 0.052 |
6.765 ± 0.089 | 6.667 ± 0.097 |
5.28 ± 0.123 | 7.013 ± 0.047 |
1.721 ± 0.032 | 1.808 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
