
Bombyx mori densovirus 3 (isolate Silkworm/China/Zhenjiang/-) (BmDNV-3)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Mouviricetes; Polivirales; Bidnaviridae; Bidensovirus; Bombyx mori bidensovirus; Bombyx mori densovirus 3
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
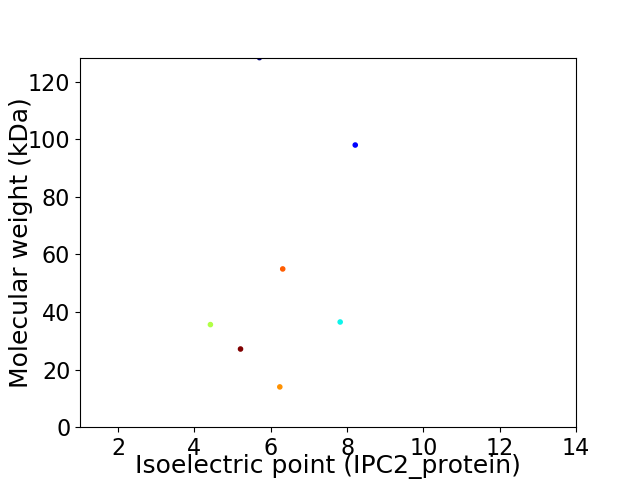
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
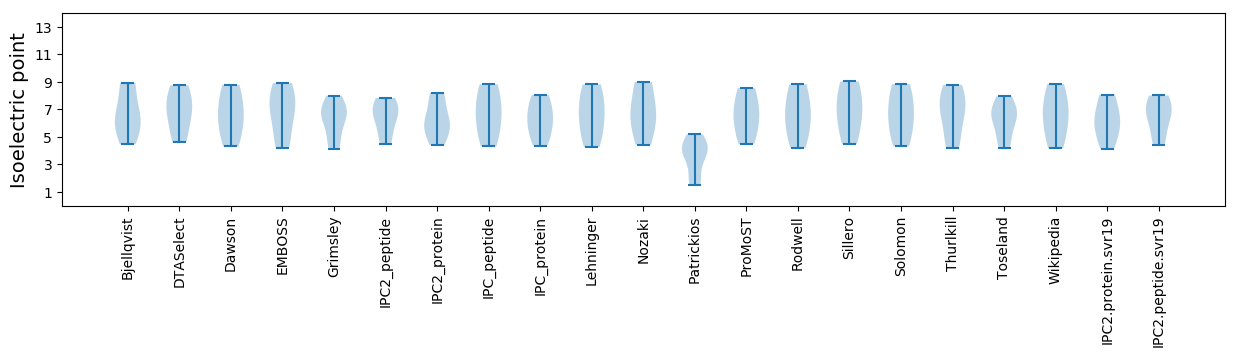
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q4U3C3|Q4U3C3_BMDND DNA-directed DNA polymerase OS=Bombyx mori densovirus 3 (isolate Silkworm/China/Zhenjiang/-) OX=648412 PE=4 SV=1
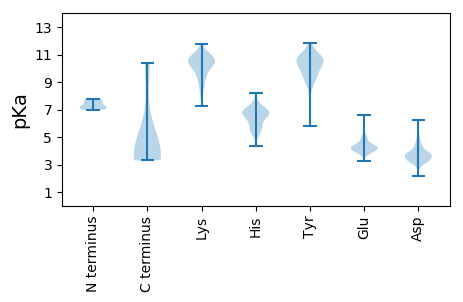
MM1 pKa = 7.16NLKK4 pKa = 10.07RR5 pKa = 11.84LYY7 pKa = 10.04VFCAVITAINSLSQSDD23 pKa = 4.85LIQIRR28 pKa = 11.84EE29 pKa = 4.3LIEE32 pKa = 3.88TNIIRR37 pKa = 11.84LSKK40 pKa = 9.35TFNGDD45 pKa = 2.16ISTINNFYY53 pKa = 10.53RR54 pKa = 11.84ILVSVFEE61 pKa = 4.87LFNDD65 pKa = 4.18LLVEE69 pKa = 4.64LDD71 pKa = 3.74TPAVSNQLQLFEE83 pKa = 6.6DD84 pKa = 4.77EE85 pKa = 4.53DD86 pKa = 4.66TVPNDD91 pKa = 3.25CLGIVGQMLIEE102 pKa = 4.17YY103 pKa = 9.79TDD105 pKa = 3.48IPNDD109 pKa = 3.61GSLTYY114 pKa = 10.96LNIGTISSLTVTLDD128 pKa = 3.24NQLYY132 pKa = 10.76LILTVIYY139 pKa = 10.38NNGRR143 pKa = 11.84EE144 pKa = 4.37TIQEE148 pKa = 3.96VHH150 pKa = 5.45NTGSPTIVVDD160 pKa = 4.62FYY162 pKa = 11.21EE163 pKa = 4.45IKK165 pKa = 10.71HH166 pKa = 6.04NNTFQEE172 pKa = 4.26IKK174 pKa = 9.79TSIYY178 pKa = 9.53YY179 pKa = 10.5VNFINFVLEE188 pKa = 4.24YY189 pKa = 8.77TVHH192 pKa = 6.65EE193 pKa = 4.36IEE195 pKa = 5.95DD196 pKa = 3.93IIVTRR201 pKa = 11.84NINRR205 pKa = 11.84YY206 pKa = 7.18NQWSTQQRR214 pKa = 11.84SRR216 pKa = 11.84SRR218 pKa = 11.84IAFIQQEE225 pKa = 4.56AIDD228 pKa = 3.83PDD230 pKa = 3.43IRR232 pKa = 11.84NEE234 pKa = 3.84QLQEE238 pKa = 3.91SAILAKK244 pKa = 10.74LHH246 pKa = 6.84DD247 pKa = 4.44AADD250 pKa = 3.57TLEE253 pKa = 4.17YY254 pKa = 10.95YY255 pKa = 10.7GISFLRR261 pKa = 11.84EE262 pKa = 3.22YY263 pKa = 10.67YY264 pKa = 9.56IAYY267 pKa = 8.94FQYY270 pKa = 10.42TIYY273 pKa = 11.21AEE275 pKa = 4.28TQKK278 pKa = 10.92LDD280 pKa = 3.26PEE282 pKa = 5.01FYY284 pKa = 10.64DD285 pKa = 5.86DD286 pKa = 4.31PLEE289 pKa = 4.62KK290 pKa = 10.62NVIYY294 pKa = 10.45INRR297 pKa = 11.84IVQALWRR304 pKa = 11.84QQ305 pKa = 3.3
MM1 pKa = 7.16NLKK4 pKa = 10.07RR5 pKa = 11.84LYY7 pKa = 10.04VFCAVITAINSLSQSDD23 pKa = 4.85LIQIRR28 pKa = 11.84EE29 pKa = 4.3LIEE32 pKa = 3.88TNIIRR37 pKa = 11.84LSKK40 pKa = 9.35TFNGDD45 pKa = 2.16ISTINNFYY53 pKa = 10.53RR54 pKa = 11.84ILVSVFEE61 pKa = 4.87LFNDD65 pKa = 4.18LLVEE69 pKa = 4.64LDD71 pKa = 3.74TPAVSNQLQLFEE83 pKa = 6.6DD84 pKa = 4.77EE85 pKa = 4.53DD86 pKa = 4.66TVPNDD91 pKa = 3.25CLGIVGQMLIEE102 pKa = 4.17YY103 pKa = 9.79TDD105 pKa = 3.48IPNDD109 pKa = 3.61GSLTYY114 pKa = 10.96LNIGTISSLTVTLDD128 pKa = 3.24NQLYY132 pKa = 10.76LILTVIYY139 pKa = 10.38NNGRR143 pKa = 11.84EE144 pKa = 4.37TIQEE148 pKa = 3.96VHH150 pKa = 5.45NTGSPTIVVDD160 pKa = 4.62FYY162 pKa = 11.21EE163 pKa = 4.45IKK165 pKa = 10.71HH166 pKa = 6.04NNTFQEE172 pKa = 4.26IKK174 pKa = 9.79TSIYY178 pKa = 9.53YY179 pKa = 10.5VNFINFVLEE188 pKa = 4.24YY189 pKa = 8.77TVHH192 pKa = 6.65EE193 pKa = 4.36IEE195 pKa = 5.95DD196 pKa = 3.93IIVTRR201 pKa = 11.84NINRR205 pKa = 11.84YY206 pKa = 7.18NQWSTQQRR214 pKa = 11.84SRR216 pKa = 11.84SRR218 pKa = 11.84IAFIQQEE225 pKa = 4.56AIDD228 pKa = 3.83PDD230 pKa = 3.43IRR232 pKa = 11.84NEE234 pKa = 3.84QLQEE238 pKa = 3.91SAILAKK244 pKa = 10.74LHH246 pKa = 6.84DD247 pKa = 4.44AADD250 pKa = 3.57TLEE253 pKa = 4.17YY254 pKa = 10.95YY255 pKa = 10.7GISFLRR261 pKa = 11.84EE262 pKa = 3.22YY263 pKa = 10.67YY264 pKa = 9.56IAYY267 pKa = 8.94FQYY270 pKa = 10.42TIYY273 pKa = 11.21AEE275 pKa = 4.28TQKK278 pKa = 10.92LDD280 pKa = 3.26PEE282 pKa = 5.01FYY284 pKa = 10.64DD285 pKa = 5.86DD286 pKa = 4.31PLEE289 pKa = 4.62KK290 pKa = 10.62NVIYY294 pKa = 10.45INRR297 pKa = 11.84IVQALWRR304 pKa = 11.84QQ305 pKa = 3.3
Molecular weight: 35.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q4U3C2|Q4U3C2_BMDND Nonstructural protein OS=Bombyx mori densovirus 3 (isolate Silkworm/China/Zhenjiang/-) OX=648412 PE=4 SV=1
MM1 pKa = 6.99EE2 pKa = 4.47TVEE5 pKa = 4.1LAPRR9 pKa = 11.84YY10 pKa = 9.0AQEE13 pKa = 4.41AKK15 pKa = 10.5PEE17 pKa = 4.05GLLYY21 pKa = 10.23PSLVQDD27 pKa = 4.18LFVQFRR33 pKa = 11.84PFIRR37 pKa = 11.84GSIPSGAGQLNPISTTVQCITRR59 pKa = 11.84GPTSAVGSLNCVQNPLQTVYY79 pKa = 10.29TYY81 pKa = 10.41PQDD84 pKa = 3.54LRR86 pKa = 11.84TIDD89 pKa = 3.68VGDD92 pKa = 3.26ITINLEE98 pKa = 3.71PNQYY102 pKa = 10.23IILRR106 pKa = 11.84LRR108 pKa = 11.84IISTGNCNFQWTVRR122 pKa = 11.84QLNINFFCSSCSGPNLICLEE142 pKa = 3.97QNNRR146 pKa = 11.84YY147 pKa = 9.0NNRR150 pKa = 11.84GSALNDD156 pKa = 3.09YY157 pKa = 10.54SFDD160 pKa = 3.58MNNIEE165 pKa = 5.16VYY167 pKa = 10.17DD168 pKa = 3.83AYY170 pKa = 11.13YY171 pKa = 10.46VNEE174 pKa = 5.16GISSAIYY181 pKa = 9.97DD182 pKa = 4.38LNPTGQLIINTRR194 pKa = 11.84LRR196 pKa = 11.84QRR198 pKa = 11.84LFVLQTPSVAEE209 pKa = 3.81YY210 pKa = 9.19TINGITRR217 pKa = 11.84PVIKK221 pKa = 10.46FNNIQFNLGTIFLSNRR237 pKa = 11.84DD238 pKa = 2.86ICATFVQTTRR248 pKa = 11.84LVIPRR253 pKa = 11.84CDD255 pKa = 3.58NAYY258 pKa = 10.47DD259 pKa = 3.34WNYY262 pKa = 10.59LFNIPGNSLATGTGVKK278 pKa = 9.67VCNNLPFRR286 pKa = 11.84FGVQRR291 pKa = 11.84CPWGPSALLPRR302 pKa = 11.84YY303 pKa = 9.3NFAWMNIIDD312 pKa = 4.22QLQEE316 pKa = 4.04LNMFNVTSFQYY327 pKa = 10.76INFRR331 pKa = 11.84YY332 pKa = 9.7NYY334 pKa = 7.32YY335 pKa = 9.01TEE337 pKa = 6.25RR338 pKa = 11.84ILTNAPIASYY348 pKa = 11.24NSAIDD353 pKa = 4.97DD354 pKa = 4.25LAQLIIDD361 pKa = 3.89EE362 pKa = 4.55GNRR365 pKa = 11.84LDD367 pKa = 4.01ALEE370 pKa = 4.63DD371 pKa = 3.21RR372 pKa = 11.84VEE374 pKa = 4.42RR375 pKa = 11.84IEE377 pKa = 5.01DD378 pKa = 3.8YY379 pKa = 10.6IIEE382 pKa = 4.19SSKK385 pKa = 8.96ITIWDD390 pKa = 3.61VLNIATNLYY399 pKa = 10.03DD400 pKa = 3.18VAGIIRR406 pKa = 11.84DD407 pKa = 3.62SAKK410 pKa = 10.75LIRR413 pKa = 11.84TASNQACKK421 pKa = 10.44SLRR424 pKa = 11.84GFFKK428 pKa = 10.75RR429 pKa = 11.84NNVAFDD435 pKa = 3.42IGNKK439 pKa = 8.96IDD441 pKa = 3.29SLTRR445 pKa = 11.84VTRR448 pKa = 11.84NNQFNPVEE456 pKa = 4.44AINYY460 pKa = 8.5GSIYY464 pKa = 9.83DD465 pKa = 4.13TIHH468 pKa = 6.57GFRR471 pKa = 11.84KK472 pKa = 8.5TSISNPTIRR481 pKa = 11.84NSNLDD486 pKa = 3.66NIITPRR492 pKa = 11.84VLHH495 pKa = 6.92DD496 pKa = 4.04NNFNLPSTSLDD507 pKa = 3.35VKK509 pKa = 11.08DD510 pKa = 5.72LINTNNVISMTTQKK524 pKa = 11.19SFMDD528 pKa = 3.86RR529 pKa = 11.84LRR531 pKa = 11.84DD532 pKa = 3.69PKK534 pKa = 10.64TGTNGDD540 pKa = 3.21IMPYY544 pKa = 10.49GVVEE548 pKa = 4.26TLASPIEE555 pKa = 4.05QLGPLGKK562 pKa = 10.09FGYY565 pKa = 10.23NRR567 pKa = 11.84DD568 pKa = 3.03ISNPSLSNFNTNSKK582 pKa = 8.69STYY585 pKa = 9.86LFHH588 pKa = 6.91TSVRR592 pKa = 11.84TKK594 pKa = 10.83SFEE597 pKa = 4.46LDD599 pKa = 3.34INNKK603 pKa = 7.77WVFSTRR609 pKa = 11.84YY610 pKa = 9.49SGIGEE615 pKa = 4.16PSVLGATTMPHH626 pKa = 5.59QIKK629 pKa = 10.45VGYY632 pKa = 10.19LKK634 pKa = 10.16IEE636 pKa = 4.23YY637 pKa = 9.41DD638 pKa = 3.85LKK640 pKa = 11.11AGTGNTVSLTRR651 pKa = 11.84RR652 pKa = 11.84NWNQVYY658 pKa = 10.54KK659 pKa = 10.85NDD661 pKa = 3.38VEE663 pKa = 4.84TYY665 pKa = 9.83TEE667 pKa = 4.23LDD669 pKa = 2.89IDD671 pKa = 4.05NLFKK675 pKa = 11.01AFSSSKK681 pKa = 9.6TYY683 pKa = 10.48RR684 pKa = 11.84EE685 pKa = 4.05RR686 pKa = 11.84QNEE689 pKa = 4.24STDD692 pKa = 3.43WKK694 pKa = 9.6WLYY697 pKa = 10.46ISFAAEE703 pKa = 3.33QKK705 pKa = 8.9YY706 pKa = 8.2NQRR709 pKa = 11.84KK710 pKa = 9.14VIQTQPIANPLYY722 pKa = 10.51LKK724 pKa = 10.9FMDD727 pKa = 4.54RR728 pKa = 11.84LFDD731 pKa = 3.77INFRR735 pKa = 11.84DD736 pKa = 3.86GRR738 pKa = 11.84LSGFKK743 pKa = 10.89YY744 pKa = 10.67NLLNQNCQTFAKK756 pKa = 8.99TQSQLLTRR764 pKa = 11.84GITNVQINDD773 pKa = 3.28QGLLGFLEE781 pKa = 4.48NYY783 pKa = 9.11KK784 pKa = 10.55QIARR788 pKa = 11.84RR789 pKa = 11.84EE790 pKa = 4.03FSLSSGMTEE799 pKa = 4.12TSWGSTSFVVRR810 pKa = 11.84ALEE813 pKa = 3.91YY814 pKa = 10.46SKK816 pKa = 11.79SNICYY821 pKa = 8.74YY822 pKa = 11.31AKK824 pKa = 10.23INNMSSNIKK833 pKa = 9.54IKK835 pKa = 10.43KK836 pKa = 8.49NRR838 pKa = 11.84YY839 pKa = 8.9SILDD843 pKa = 3.47KK844 pKa = 10.8AYY846 pKa = 10.02KK847 pKa = 9.86NDD849 pKa = 3.18NTIRR853 pKa = 11.84TVICTT858 pKa = 3.58
MM1 pKa = 6.99EE2 pKa = 4.47TVEE5 pKa = 4.1LAPRR9 pKa = 11.84YY10 pKa = 9.0AQEE13 pKa = 4.41AKK15 pKa = 10.5PEE17 pKa = 4.05GLLYY21 pKa = 10.23PSLVQDD27 pKa = 4.18LFVQFRR33 pKa = 11.84PFIRR37 pKa = 11.84GSIPSGAGQLNPISTTVQCITRR59 pKa = 11.84GPTSAVGSLNCVQNPLQTVYY79 pKa = 10.29TYY81 pKa = 10.41PQDD84 pKa = 3.54LRR86 pKa = 11.84TIDD89 pKa = 3.68VGDD92 pKa = 3.26ITINLEE98 pKa = 3.71PNQYY102 pKa = 10.23IILRR106 pKa = 11.84LRR108 pKa = 11.84IISTGNCNFQWTVRR122 pKa = 11.84QLNINFFCSSCSGPNLICLEE142 pKa = 3.97QNNRR146 pKa = 11.84YY147 pKa = 9.0NNRR150 pKa = 11.84GSALNDD156 pKa = 3.09YY157 pKa = 10.54SFDD160 pKa = 3.58MNNIEE165 pKa = 5.16VYY167 pKa = 10.17DD168 pKa = 3.83AYY170 pKa = 11.13YY171 pKa = 10.46VNEE174 pKa = 5.16GISSAIYY181 pKa = 9.97DD182 pKa = 4.38LNPTGQLIINTRR194 pKa = 11.84LRR196 pKa = 11.84QRR198 pKa = 11.84LFVLQTPSVAEE209 pKa = 3.81YY210 pKa = 9.19TINGITRR217 pKa = 11.84PVIKK221 pKa = 10.46FNNIQFNLGTIFLSNRR237 pKa = 11.84DD238 pKa = 2.86ICATFVQTTRR248 pKa = 11.84LVIPRR253 pKa = 11.84CDD255 pKa = 3.58NAYY258 pKa = 10.47DD259 pKa = 3.34WNYY262 pKa = 10.59LFNIPGNSLATGTGVKK278 pKa = 9.67VCNNLPFRR286 pKa = 11.84FGVQRR291 pKa = 11.84CPWGPSALLPRR302 pKa = 11.84YY303 pKa = 9.3NFAWMNIIDD312 pKa = 4.22QLQEE316 pKa = 4.04LNMFNVTSFQYY327 pKa = 10.76INFRR331 pKa = 11.84YY332 pKa = 9.7NYY334 pKa = 7.32YY335 pKa = 9.01TEE337 pKa = 6.25RR338 pKa = 11.84ILTNAPIASYY348 pKa = 11.24NSAIDD353 pKa = 4.97DD354 pKa = 4.25LAQLIIDD361 pKa = 3.89EE362 pKa = 4.55GNRR365 pKa = 11.84LDD367 pKa = 4.01ALEE370 pKa = 4.63DD371 pKa = 3.21RR372 pKa = 11.84VEE374 pKa = 4.42RR375 pKa = 11.84IEE377 pKa = 5.01DD378 pKa = 3.8YY379 pKa = 10.6IIEE382 pKa = 4.19SSKK385 pKa = 8.96ITIWDD390 pKa = 3.61VLNIATNLYY399 pKa = 10.03DD400 pKa = 3.18VAGIIRR406 pKa = 11.84DD407 pKa = 3.62SAKK410 pKa = 10.75LIRR413 pKa = 11.84TASNQACKK421 pKa = 10.44SLRR424 pKa = 11.84GFFKK428 pKa = 10.75RR429 pKa = 11.84NNVAFDD435 pKa = 3.42IGNKK439 pKa = 8.96IDD441 pKa = 3.29SLTRR445 pKa = 11.84VTRR448 pKa = 11.84NNQFNPVEE456 pKa = 4.44AINYY460 pKa = 8.5GSIYY464 pKa = 9.83DD465 pKa = 4.13TIHH468 pKa = 6.57GFRR471 pKa = 11.84KK472 pKa = 8.5TSISNPTIRR481 pKa = 11.84NSNLDD486 pKa = 3.66NIITPRR492 pKa = 11.84VLHH495 pKa = 6.92DD496 pKa = 4.04NNFNLPSTSLDD507 pKa = 3.35VKK509 pKa = 11.08DD510 pKa = 5.72LINTNNVISMTTQKK524 pKa = 11.19SFMDD528 pKa = 3.86RR529 pKa = 11.84LRR531 pKa = 11.84DD532 pKa = 3.69PKK534 pKa = 10.64TGTNGDD540 pKa = 3.21IMPYY544 pKa = 10.49GVVEE548 pKa = 4.26TLASPIEE555 pKa = 4.05QLGPLGKK562 pKa = 10.09FGYY565 pKa = 10.23NRR567 pKa = 11.84DD568 pKa = 3.03ISNPSLSNFNTNSKK582 pKa = 8.69STYY585 pKa = 9.86LFHH588 pKa = 6.91TSVRR592 pKa = 11.84TKK594 pKa = 10.83SFEE597 pKa = 4.46LDD599 pKa = 3.34INNKK603 pKa = 7.77WVFSTRR609 pKa = 11.84YY610 pKa = 9.49SGIGEE615 pKa = 4.16PSVLGATTMPHH626 pKa = 5.59QIKK629 pKa = 10.45VGYY632 pKa = 10.19LKK634 pKa = 10.16IEE636 pKa = 4.23YY637 pKa = 9.41DD638 pKa = 3.85LKK640 pKa = 11.11AGTGNTVSLTRR651 pKa = 11.84RR652 pKa = 11.84NWNQVYY658 pKa = 10.54KK659 pKa = 10.85NDD661 pKa = 3.38VEE663 pKa = 4.84TYY665 pKa = 9.83TEE667 pKa = 4.23LDD669 pKa = 2.89IDD671 pKa = 4.05NLFKK675 pKa = 11.01AFSSSKK681 pKa = 9.6TYY683 pKa = 10.48RR684 pKa = 11.84EE685 pKa = 4.05RR686 pKa = 11.84QNEE689 pKa = 4.24STDD692 pKa = 3.43WKK694 pKa = 9.6WLYY697 pKa = 10.46ISFAAEE703 pKa = 3.33QKK705 pKa = 8.9YY706 pKa = 8.2NQRR709 pKa = 11.84KK710 pKa = 9.14VIQTQPIANPLYY722 pKa = 10.51LKK724 pKa = 10.9FMDD727 pKa = 4.54RR728 pKa = 11.84LFDD731 pKa = 3.77INFRR735 pKa = 11.84DD736 pKa = 3.86GRR738 pKa = 11.84LSGFKK743 pKa = 10.89YY744 pKa = 10.67NLLNQNCQTFAKK756 pKa = 8.99TQSQLLTRR764 pKa = 11.84GITNVQINDD773 pKa = 3.28QGLLGFLEE781 pKa = 4.48NYY783 pKa = 9.11KK784 pKa = 10.55QIARR788 pKa = 11.84RR789 pKa = 11.84EE790 pKa = 4.03FSLSSGMTEE799 pKa = 4.12TSWGSTSFVVRR810 pKa = 11.84ALEE813 pKa = 3.91YY814 pKa = 10.46SKK816 pKa = 11.79SNICYY821 pKa = 8.74YY822 pKa = 11.31AKK824 pKa = 10.23INNMSSNIKK833 pKa = 9.54IKK835 pKa = 10.43KK836 pKa = 8.49NRR838 pKa = 11.84YY839 pKa = 8.9SILDD843 pKa = 3.47KK844 pKa = 10.8AYY846 pKa = 10.02KK847 pKa = 9.86NDD849 pKa = 3.18NTIRR853 pKa = 11.84TVICTT858 pKa = 3.58
Molecular weight: 97.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3441 |
126 |
1115 |
491.6 |
56.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.359 ± 0.486 | 1.715 ± 0.309 |
6.132 ± 0.609 | 5.551 ± 0.595 |
5.086 ± 0.387 | 5.173 ± 0.819 |
1.453 ± 0.384 | 8.66 ± 0.507 |
6.452 ± 1.132 | 8.37 ± 0.529 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.598 ± 0.186 | 8.166 ± 0.714 |
4.098 ± 0.236 | 3.226 ± 0.64 |
4.621 ± 0.723 | 7.469 ± 0.784 |
6.423 ± 0.448 | 4.911 ± 0.308 |
1.104 ± 0.247 | 5.434 ± 0.478 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
