
Roseobacter phage CRP-3
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; unclassified Podoviridae
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
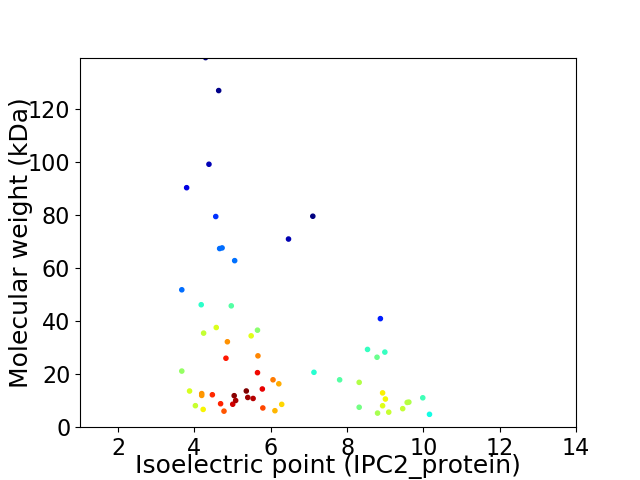
Virtual 2D-PAGE plot for 60 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
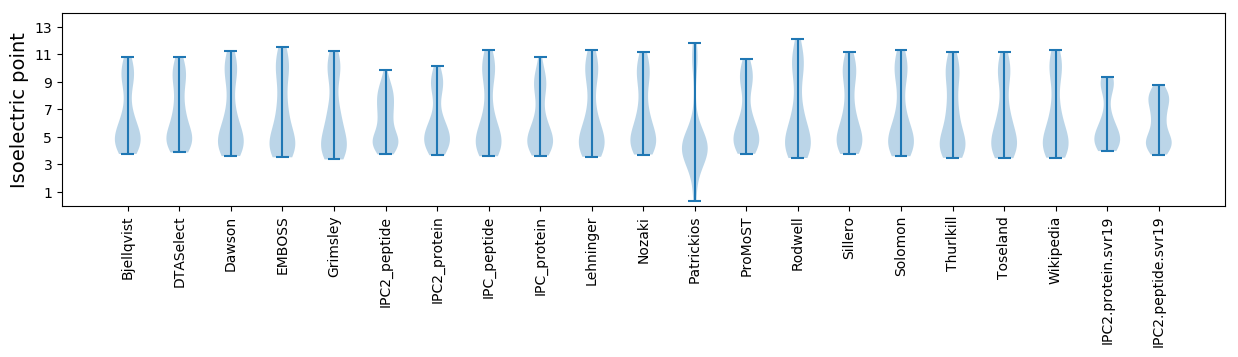
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A646QVZ6|A0A646QVZ6_9CAUD Uncharacterized protein OS=Roseobacter phage CRP-3 OX=2559282 GN=CRP3_gp46 PE=4 SV=1
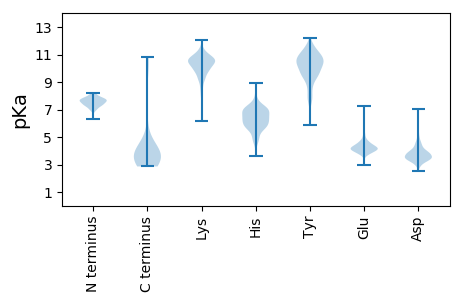
MM1 pKa = 7.64AGYY4 pKa = 7.86TRR6 pKa = 11.84QSTSQIINGANITAPPLNAEE26 pKa = 4.47FNQLASSFGVTGHH39 pKa = 5.4THH41 pKa = 7.57DD42 pKa = 4.42GTSGNAPKK50 pKa = 9.83IDD52 pKa = 3.77LATSVTGYY60 pKa = 10.97LPATNGGTGGKK71 pKa = 9.71NNLAATTNPGNGDD84 pKa = 3.92DD85 pKa = 5.43ADD87 pKa = 4.35DD88 pKa = 4.92GYY90 pKa = 11.99SRR92 pKa = 11.84GSYY95 pKa = 8.01WYY97 pKa = 10.17NYY99 pKa = 9.25TGDD102 pKa = 3.11RR103 pKa = 11.84WYY105 pKa = 10.09ICINNTVGSAVWRR118 pKa = 11.84EE119 pKa = 3.77MLMVEE124 pKa = 4.35TGSVIEE130 pKa = 4.37PGITGAVDD138 pKa = 3.96LGSTTFKK145 pKa = 11.01FKK147 pKa = 9.73DD148 pKa = 3.34THH150 pKa = 7.28LSGSMNSVSAAISGNITVGGTQTNTGAATFNGSATFNNITNFNGASNTIANAAITSGTINGTTIGGATAAAGTFTALTANNSANLTNATISGGTINNSVIGGTTPLNITGLTVAASTGFSGDD272 pKa = 2.95LTGNVLGNVTGNLTGTVNATGGVIGDD298 pKa = 3.4VTGNVTASSGSSTFNNVVINGSLDD322 pKa = 3.25MDD324 pKa = 3.85ATTASTITGLSTPVNPTDD342 pKa = 3.94ASTKK346 pKa = 10.44AYY348 pKa = 10.69VDD350 pKa = 3.42QQVALVLSSAPAALDD365 pKa = 3.51TLNEE369 pKa = 3.77LAAAINDD376 pKa = 3.85DD377 pKa = 4.31ANFATTVNNSIATKK391 pKa = 10.36LSLSGGTMSGNIDD404 pKa = 3.74LNSTNKK410 pKa = 7.44ITNMPTPSANSDD422 pKa = 3.44GSNKK426 pKa = 10.13GYY428 pKa = 11.0VDD430 pKa = 3.49TQRR433 pKa = 11.84DD434 pKa = 3.48TRR436 pKa = 11.84LATTGGTMSGAINMNSQAITSLANPANSGDD466 pKa = 3.83AANKK470 pKa = 10.02FYY472 pKa = 11.14VDD474 pKa = 4.79SILGSATSAAASASTASALAAQASGSAANAQASEE508 pKa = 4.73DD509 pKa = 4.12EE510 pKa = 4.27AQEE513 pKa = 4.03WATKK517 pKa = 6.57TTGTITGEE525 pKa = 4.03SEE527 pKa = 4.16YY528 pKa = 10.83SAKK531 pKa = 10.18EE532 pKa = 3.8YY533 pKa = 10.79AIGTVIRR540 pKa = 11.84GNIGSAKK547 pKa = 10.2DD548 pKa = 2.96WSTYY552 pKa = 9.59TGGTVDD558 pKa = 3.57GTNYY562 pKa = 9.26SAKK565 pKa = 10.26YY566 pKa = 8.05WATNANIGTIASNIDD581 pKa = 3.45DD582 pKa = 4.62LVNVANDD589 pKa = 3.91LSSGNFVAGQIYY601 pKa = 10.37DD602 pKa = 3.72FGSITDD608 pKa = 3.9AASGTSGAPDD618 pKa = 3.55GFIVTVANNLADD630 pKa = 3.74VQTVSNVITNVNTVAGISANVTTVAGVSSAVSTLAAISSDD670 pKa = 3.38VSAVAPIAANVTTVAGVSSDD690 pKa = 3.42VTAVANNNANINTVVGQITPNNNISTLASIAGNISTVASVSVDD733 pKa = 3.21VTTVAGISSNVTAVANNNANVTSVANIDD761 pKa = 3.62SDD763 pKa = 4.0VTAVAGIASDD773 pKa = 3.68VTTAATNVTDD783 pKa = 3.33ITNFADD789 pKa = 4.38VYY791 pKa = 10.5QGPKK795 pKa = 8.55ATAPTQRR802 pKa = 11.84NNNSSLVAGDD812 pKa = 4.72LYY814 pKa = 11.31FDD816 pKa = 3.91TVSGFMRR823 pKa = 11.84YY824 pKa = 9.11YY825 pKa = 10.78SGSSWSNITAPTGDD839 pKa = 3.8MGNQNSTSVAITGGSITGITDD860 pKa = 3.8LEE862 pKa = 4.37IADD865 pKa = 4.53GGTGASSAPAARR877 pKa = 11.84TNLGLDD883 pKa = 3.51TMATQAANNVAITGGTISVDD903 pKa = 3.79FDD905 pKa = 3.9FGSII909 pKa = 3.53
MM1 pKa = 7.64AGYY4 pKa = 7.86TRR6 pKa = 11.84QSTSQIINGANITAPPLNAEE26 pKa = 4.47FNQLASSFGVTGHH39 pKa = 5.4THH41 pKa = 7.57DD42 pKa = 4.42GTSGNAPKK50 pKa = 9.83IDD52 pKa = 3.77LATSVTGYY60 pKa = 10.97LPATNGGTGGKK71 pKa = 9.71NNLAATTNPGNGDD84 pKa = 3.92DD85 pKa = 5.43ADD87 pKa = 4.35DD88 pKa = 4.92GYY90 pKa = 11.99SRR92 pKa = 11.84GSYY95 pKa = 8.01WYY97 pKa = 10.17NYY99 pKa = 9.25TGDD102 pKa = 3.11RR103 pKa = 11.84WYY105 pKa = 10.09ICINNTVGSAVWRR118 pKa = 11.84EE119 pKa = 3.77MLMVEE124 pKa = 4.35TGSVIEE130 pKa = 4.37PGITGAVDD138 pKa = 3.96LGSTTFKK145 pKa = 11.01FKK147 pKa = 9.73DD148 pKa = 3.34THH150 pKa = 7.28LSGSMNSVSAAISGNITVGGTQTNTGAATFNGSATFNNITNFNGASNTIANAAITSGTINGTTIGGATAAAGTFTALTANNSANLTNATISGGTINNSVIGGTTPLNITGLTVAASTGFSGDD272 pKa = 2.95LTGNVLGNVTGNLTGTVNATGGVIGDD298 pKa = 3.4VTGNVTASSGSSTFNNVVINGSLDD322 pKa = 3.25MDD324 pKa = 3.85ATTASTITGLSTPVNPTDD342 pKa = 3.94ASTKK346 pKa = 10.44AYY348 pKa = 10.69VDD350 pKa = 3.42QQVALVLSSAPAALDD365 pKa = 3.51TLNEE369 pKa = 3.77LAAAINDD376 pKa = 3.85DD377 pKa = 4.31ANFATTVNNSIATKK391 pKa = 10.36LSLSGGTMSGNIDD404 pKa = 3.74LNSTNKK410 pKa = 7.44ITNMPTPSANSDD422 pKa = 3.44GSNKK426 pKa = 10.13GYY428 pKa = 11.0VDD430 pKa = 3.49TQRR433 pKa = 11.84DD434 pKa = 3.48TRR436 pKa = 11.84LATTGGTMSGAINMNSQAITSLANPANSGDD466 pKa = 3.83AANKK470 pKa = 10.02FYY472 pKa = 11.14VDD474 pKa = 4.79SILGSATSAAASASTASALAAQASGSAANAQASEE508 pKa = 4.73DD509 pKa = 4.12EE510 pKa = 4.27AQEE513 pKa = 4.03WATKK517 pKa = 6.57TTGTITGEE525 pKa = 4.03SEE527 pKa = 4.16YY528 pKa = 10.83SAKK531 pKa = 10.18EE532 pKa = 3.8YY533 pKa = 10.79AIGTVIRR540 pKa = 11.84GNIGSAKK547 pKa = 10.2DD548 pKa = 2.96WSTYY552 pKa = 9.59TGGTVDD558 pKa = 3.57GTNYY562 pKa = 9.26SAKK565 pKa = 10.26YY566 pKa = 8.05WATNANIGTIASNIDD581 pKa = 3.45DD582 pKa = 4.62LVNVANDD589 pKa = 3.91LSSGNFVAGQIYY601 pKa = 10.37DD602 pKa = 3.72FGSITDD608 pKa = 3.9AASGTSGAPDD618 pKa = 3.55GFIVTVANNLADD630 pKa = 3.74VQTVSNVITNVNTVAGISANVTTVAGVSSAVSTLAAISSDD670 pKa = 3.38VSAVAPIAANVTTVAGVSSDD690 pKa = 3.42VTAVANNNANINTVVGQITPNNNISTLASIAGNISTVASVSVDD733 pKa = 3.21VTTVAGISSNVTAVANNNANVTSVANIDD761 pKa = 3.62SDD763 pKa = 4.0VTAVAGIASDD773 pKa = 3.68VTTAATNVTDD783 pKa = 3.33ITNFADD789 pKa = 4.38VYY791 pKa = 10.5QGPKK795 pKa = 8.55ATAPTQRR802 pKa = 11.84NNNSSLVAGDD812 pKa = 4.72LYY814 pKa = 11.31FDD816 pKa = 3.91TVSGFMRR823 pKa = 11.84YY824 pKa = 9.11YY825 pKa = 10.78SGSSWSNITAPTGDD839 pKa = 3.8MGNQNSTSVAITGGSITGITDD860 pKa = 3.8LEE862 pKa = 4.37IADD865 pKa = 4.53GGTGASSAPAARR877 pKa = 11.84TNLGLDD883 pKa = 3.51TMATQAANNVAITGGTISVDD903 pKa = 3.79FDD905 pKa = 3.9FGSII909 pKa = 3.53
Molecular weight: 90.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A646QX19|A0A646QX19_9CAUD Uncharacterized protein OS=Roseobacter phage CRP-3 OX=2559282 GN=CRP3_gp03 PE=4 SV=1
MM1 pKa = 7.48YY2 pKa = 10.45ILGANFLLAGQLKK15 pKa = 9.48IRR17 pKa = 11.84AALRR21 pKa = 11.84GGFFLTAPRR30 pKa = 11.84AIFRR34 pKa = 11.84RR35 pKa = 11.84FEE37 pKa = 3.86WGVIGPGILTQKK49 pKa = 10.56NPFPVFSGDD58 pKa = 3.28LVFFRR63 pKa = 11.84VYY65 pKa = 10.44FMAFCTVFFTPLYY78 pKa = 9.23IVRR81 pKa = 11.84SIHH84 pKa = 6.13NLGKK88 pKa = 10.43VFPHH92 pKa = 7.43LIKK95 pKa = 10.92CLL97 pKa = 3.55
MM1 pKa = 7.48YY2 pKa = 10.45ILGANFLLAGQLKK15 pKa = 9.48IRR17 pKa = 11.84AALRR21 pKa = 11.84GGFFLTAPRR30 pKa = 11.84AIFRR34 pKa = 11.84RR35 pKa = 11.84FEE37 pKa = 3.86WGVIGPGILTQKK49 pKa = 10.56NPFPVFSGDD58 pKa = 3.28LVFFRR63 pKa = 11.84VYY65 pKa = 10.44FMAFCTVFFTPLYY78 pKa = 9.23IVRR81 pKa = 11.84SIHH84 pKa = 6.13NLGKK88 pKa = 10.43VFPHH92 pKa = 7.43LIKK95 pKa = 10.92CLL97 pKa = 3.55
Molecular weight: 11.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
16287 |
43 |
1292 |
271.4 |
29.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.603 ± 0.545 | 0.786 ± 0.128 |
6.907 ± 0.304 | 6.766 ± 0.445 |
3.764 ± 0.181 | 8.479 ± 0.589 |
1.351 ± 0.208 | 5.25 ± 0.172 |
5.391 ± 0.398 | 7.472 ± 0.303 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.935 ± 0.226 | 5.047 ± 0.336 |
3.917 ± 0.302 | 4.071 ± 0.262 |
5.004 ± 0.321 | 6.576 ± 0.319 |
6.564 ± 0.451 | 6.011 ± 0.257 |
1.068 ± 0.132 | 3.039 ± 0.2 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
