
Brevibacillus panacihumi W25
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Paenibacillaceae; Brevibacillus; Brevibacillus panacihumi
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
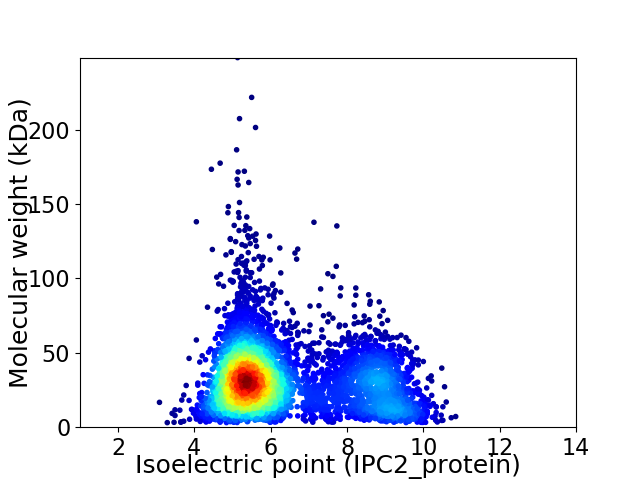
Virtual 2D-PAGE plot for 5188 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V6M4R0|V6M4R0_9BACL Spore germination protein OS=Brevibacillus panacihumi W25 OX=1408254 GN=T458_22635 PE=3 SV=1
MM1 pKa = 7.27KK2 pKa = 10.52ALVEE6 pKa = 4.81FCASNVASYY15 pKa = 8.43TQAVMDD21 pKa = 5.16ALEE24 pKa = 4.37QDD26 pKa = 3.76PEE28 pKa = 4.65LDD30 pKa = 3.43VDD32 pKa = 3.95VLEE35 pKa = 4.5YY36 pKa = 11.28GCLGYY41 pKa = 10.76CGEE44 pKa = 5.14CYY46 pKa = 10.62LEE48 pKa = 4.78PFALVNGKK56 pKa = 8.92LVQAPTAEE64 pKa = 4.13ALLDD68 pKa = 4.26KK69 pKa = 10.56IKK71 pKa = 10.74QVLKK75 pKa = 11.07EE76 pKa = 3.97EE77 pKa = 4.9DD78 pKa = 4.59DD79 pKa = 4.0PFKK82 pKa = 10.96DD83 pKa = 4.34LPLL86 pKa = 5.41
MM1 pKa = 7.27KK2 pKa = 10.52ALVEE6 pKa = 4.81FCASNVASYY15 pKa = 8.43TQAVMDD21 pKa = 5.16ALEE24 pKa = 4.37QDD26 pKa = 3.76PEE28 pKa = 4.65LDD30 pKa = 3.43VDD32 pKa = 3.95VLEE35 pKa = 4.5YY36 pKa = 11.28GCLGYY41 pKa = 10.76CGEE44 pKa = 5.14CYY46 pKa = 10.62LEE48 pKa = 4.78PFALVNGKK56 pKa = 8.92LVQAPTAEE64 pKa = 4.13ALLDD68 pKa = 4.26KK69 pKa = 10.56IKK71 pKa = 10.74QVLKK75 pKa = 11.07EE76 pKa = 3.97EE77 pKa = 4.9DD78 pKa = 4.59DD79 pKa = 4.0PFKK82 pKa = 10.96DD83 pKa = 4.34LPLL86 pKa = 5.41
Molecular weight: 9.49 kDa
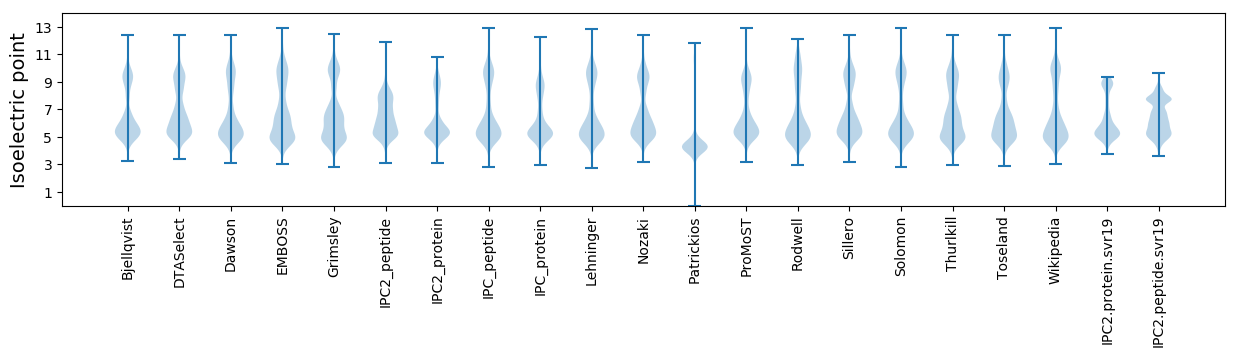
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V6MAB5|V6MAB5_9BACL Membrane protein OS=Brevibacillus panacihumi W25 OX=1408254 GN=T458_07100 PE=3 SV=1
MM1 pKa = 7.75SYY3 pKa = 11.08LPTVVQAIEE12 pKa = 4.18PKK14 pKa = 10.43SEE16 pKa = 3.67VDD18 pKa = 3.72LYY20 pKa = 11.31LPQAHH25 pKa = 5.02MRR27 pKa = 11.84KK28 pKa = 9.13WNLQPSTLTVQFGNKK43 pKa = 6.61TVRR46 pKa = 11.84ARR48 pKa = 11.84VSRR51 pKa = 11.84MEE53 pKa = 3.98RR54 pKa = 11.84KK55 pKa = 8.88SRR57 pKa = 11.84IWIRR61 pKa = 11.84TSMAQSLHH69 pKa = 6.57LPTGVPLLVDD79 pKa = 3.35YY80 pKa = 10.36QANQQSLVFGPYY92 pKa = 9.31MGILVSAYY100 pKa = 9.79NLQYY104 pKa = 10.66PLAPFGPLSHH114 pKa = 7.29FFNEE118 pKa = 4.44VADD121 pKa = 4.06SCRR124 pKa = 11.84KK125 pKa = 9.5RR126 pKa = 11.84GGVICAFRR134 pKa = 11.84PQDD137 pKa = 3.79VNWDD141 pKa = 3.39AGIVRR146 pKa = 11.84GLVRR150 pKa = 11.84KK151 pKa = 9.88GGSWRR156 pKa = 11.84QQVLPLPQCIYY167 pKa = 10.31NRR169 pKa = 11.84LVSRR173 pKa = 11.84QRR175 pKa = 11.84EE176 pKa = 3.79RR177 pKa = 11.84SEE179 pKa = 5.3AMSAWVQRR187 pKa = 11.84CKK189 pKa = 10.38EE190 pKa = 3.84ANIPFFNEE198 pKa = 3.18RR199 pKa = 11.84FLNKK203 pKa = 8.65WHH205 pKa = 6.0VHH207 pKa = 4.35TALEE211 pKa = 4.21KK212 pKa = 10.38QEE214 pKa = 4.3AAVPHH219 pKa = 6.14LPGMLRR225 pKa = 11.84YY226 pKa = 9.43EE227 pKa = 4.31NQDD230 pKa = 3.65DD231 pKa = 4.22LQEE234 pKa = 4.46MITRR238 pKa = 11.84YY239 pKa = 7.86HH240 pKa = 5.27TVYY243 pKa = 10.79AKK245 pKa = 10.03PANGSMGRR253 pKa = 11.84GIIRR257 pKa = 11.84LRR259 pKa = 11.84RR260 pKa = 11.84KK261 pKa = 9.49QNGYY265 pKa = 9.02QAASPGGLNKK275 pKa = 10.08HH276 pKa = 6.26FSSISGLHH284 pKa = 6.21KK285 pKa = 10.62YY286 pKa = 9.92LSKK289 pKa = 9.88RR290 pKa = 11.84TKK292 pKa = 9.95GKK294 pKa = 9.78PYY296 pKa = 10.54LLQQGLPLIGVQNRR310 pKa = 11.84PADD313 pKa = 3.78FRR315 pKa = 11.84VLVQKK320 pKa = 10.33DD321 pKa = 3.72RR322 pKa = 11.84KK323 pKa = 9.83GEE325 pKa = 3.75WSITSLVARR334 pKa = 11.84LGQNRR339 pKa = 11.84IVSNVSRR346 pKa = 11.84GGSMMSPAHH355 pKa = 6.95ALRR358 pKa = 11.84LCGPWACSQRR368 pKa = 11.84PTPQTLKK375 pKa = 10.74HH376 pKa = 5.52VALKK380 pKa = 10.6LSRR383 pKa = 11.84LLEE386 pKa = 4.17EE387 pKa = 4.94ALPGHH392 pKa = 5.63YY393 pKa = 10.59AEE395 pKa = 5.36FGVDD399 pKa = 3.17LGVDD403 pKa = 3.26VRR405 pKa = 11.84GQVWLLEE412 pKa = 4.49VNSKK416 pKa = 10.07PSKK419 pKa = 8.71TVNSIPIPEE428 pKa = 4.38GAEE431 pKa = 3.81APPRR435 pKa = 11.84RR436 pKa = 11.84ARR438 pKa = 11.84PSAVRR443 pKa = 11.84MFEE446 pKa = 3.93YY447 pKa = 10.61ASYY450 pKa = 10.85ISGFPRR456 pKa = 11.84PSPNRR461 pKa = 11.84KK462 pKa = 5.93TARR465 pKa = 11.84KK466 pKa = 6.76KK467 pKa = 7.74TRR469 pKa = 11.84RR470 pKa = 11.84RR471 pKa = 3.38
MM1 pKa = 7.75SYY3 pKa = 11.08LPTVVQAIEE12 pKa = 4.18PKK14 pKa = 10.43SEE16 pKa = 3.67VDD18 pKa = 3.72LYY20 pKa = 11.31LPQAHH25 pKa = 5.02MRR27 pKa = 11.84KK28 pKa = 9.13WNLQPSTLTVQFGNKK43 pKa = 6.61TVRR46 pKa = 11.84ARR48 pKa = 11.84VSRR51 pKa = 11.84MEE53 pKa = 3.98RR54 pKa = 11.84KK55 pKa = 8.88SRR57 pKa = 11.84IWIRR61 pKa = 11.84TSMAQSLHH69 pKa = 6.57LPTGVPLLVDD79 pKa = 3.35YY80 pKa = 10.36QANQQSLVFGPYY92 pKa = 9.31MGILVSAYY100 pKa = 9.79NLQYY104 pKa = 10.66PLAPFGPLSHH114 pKa = 7.29FFNEE118 pKa = 4.44VADD121 pKa = 4.06SCRR124 pKa = 11.84KK125 pKa = 9.5RR126 pKa = 11.84GGVICAFRR134 pKa = 11.84PQDD137 pKa = 3.79VNWDD141 pKa = 3.39AGIVRR146 pKa = 11.84GLVRR150 pKa = 11.84KK151 pKa = 9.88GGSWRR156 pKa = 11.84QQVLPLPQCIYY167 pKa = 10.31NRR169 pKa = 11.84LVSRR173 pKa = 11.84QRR175 pKa = 11.84EE176 pKa = 3.79RR177 pKa = 11.84SEE179 pKa = 5.3AMSAWVQRR187 pKa = 11.84CKK189 pKa = 10.38EE190 pKa = 3.84ANIPFFNEE198 pKa = 3.18RR199 pKa = 11.84FLNKK203 pKa = 8.65WHH205 pKa = 6.0VHH207 pKa = 4.35TALEE211 pKa = 4.21KK212 pKa = 10.38QEE214 pKa = 4.3AAVPHH219 pKa = 6.14LPGMLRR225 pKa = 11.84YY226 pKa = 9.43EE227 pKa = 4.31NQDD230 pKa = 3.65DD231 pKa = 4.22LQEE234 pKa = 4.46MITRR238 pKa = 11.84YY239 pKa = 7.86HH240 pKa = 5.27TVYY243 pKa = 10.79AKK245 pKa = 10.03PANGSMGRR253 pKa = 11.84GIIRR257 pKa = 11.84LRR259 pKa = 11.84RR260 pKa = 11.84KK261 pKa = 9.49QNGYY265 pKa = 9.02QAASPGGLNKK275 pKa = 10.08HH276 pKa = 6.26FSSISGLHH284 pKa = 6.21KK285 pKa = 10.62YY286 pKa = 9.92LSKK289 pKa = 9.88RR290 pKa = 11.84TKK292 pKa = 9.95GKK294 pKa = 9.78PYY296 pKa = 10.54LLQQGLPLIGVQNRR310 pKa = 11.84PADD313 pKa = 3.78FRR315 pKa = 11.84VLVQKK320 pKa = 10.33DD321 pKa = 3.72RR322 pKa = 11.84KK323 pKa = 9.83GEE325 pKa = 3.75WSITSLVARR334 pKa = 11.84LGQNRR339 pKa = 11.84IVSNVSRR346 pKa = 11.84GGSMMSPAHH355 pKa = 6.95ALRR358 pKa = 11.84LCGPWACSQRR368 pKa = 11.84PTPQTLKK375 pKa = 10.74HH376 pKa = 5.52VALKK380 pKa = 10.6LSRR383 pKa = 11.84LLEE386 pKa = 4.17EE387 pKa = 4.94ALPGHH392 pKa = 5.63YY393 pKa = 10.59AEE395 pKa = 5.36FGVDD399 pKa = 3.17LGVDD403 pKa = 3.26VRR405 pKa = 11.84GQVWLLEE412 pKa = 4.49VNSKK416 pKa = 10.07PSKK419 pKa = 8.71TVNSIPIPEE428 pKa = 4.38GAEE431 pKa = 3.81APPRR435 pKa = 11.84RR436 pKa = 11.84ARR438 pKa = 11.84PSAVRR443 pKa = 11.84MFEE446 pKa = 3.93YY447 pKa = 10.61ASYY450 pKa = 10.85ISGFPRR456 pKa = 11.84PSPNRR461 pKa = 11.84KK462 pKa = 5.93TARR465 pKa = 11.84KK466 pKa = 6.76KK467 pKa = 7.74TRR469 pKa = 11.84RR470 pKa = 11.84RR471 pKa = 3.38
Molecular weight: 53.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1544730 |
27 |
2184 |
297.8 |
33.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.254 ± 0.039 | 0.792 ± 0.01 |
4.88 ± 0.024 | 7.212 ± 0.041 |
4.064 ± 0.027 | 7.202 ± 0.033 |
2.203 ± 0.018 | 6.638 ± 0.035 |
5.483 ± 0.031 | 10.229 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.913 ± 0.02 | 3.384 ± 0.022 |
4.167 ± 0.024 | 4.32 ± 0.029 |
5.243 ± 0.032 | 5.863 ± 0.025 |
5.306 ± 0.024 | 7.394 ± 0.028 |
1.223 ± 0.013 | 3.229 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
