
Magnaporthe oryzae virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
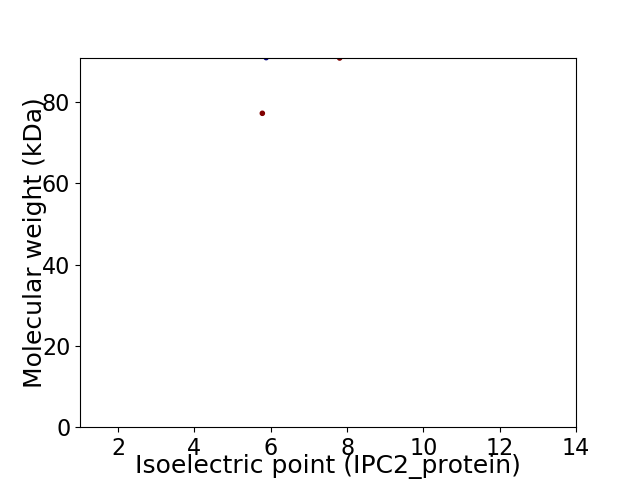
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q60GI3|Q60GI3_9VIRU Putative coat protein OS=Magnaporthe oryzae virus 1 OX=271257 PE=4 SV=1
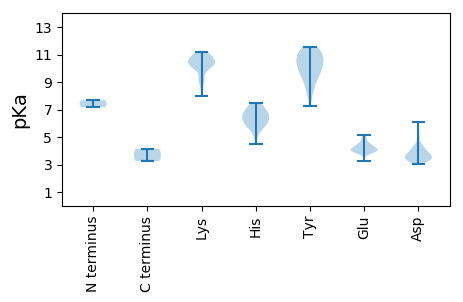
MM1 pKa = 7.18AQIGAPNFLSSVLGDD16 pKa = 3.38QRR18 pKa = 11.84GGVLNSDD25 pKa = 3.67SVFRR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 9.22RR32 pKa = 11.84AAITTSTIVRR42 pKa = 11.84GVQDD46 pKa = 3.41TRR48 pKa = 11.84VGFLVFAVGRR58 pKa = 11.84VFSSFSAALVRR69 pKa = 11.84PKK71 pKa = 10.85SSVPAIDD78 pKa = 3.32ATYY81 pKa = 8.75PCPATRR87 pKa = 11.84SEE89 pKa = 4.4EE90 pKa = 4.11FVGLAKK96 pKa = 10.32KK97 pKa = 10.47YY98 pKa = 10.76SNFSSTFTHH107 pKa = 6.79ANFAGVVEE115 pKa = 4.22RR116 pKa = 11.84LSRR119 pKa = 11.84ALAVLSTLPDD129 pKa = 3.42SDD131 pKa = 3.95TAALTSLDD139 pKa = 3.22IAGGVMPTVYY149 pKa = 10.55SIATFDD155 pKa = 3.86SPVNALARR163 pKa = 11.84VVFIPRR169 pKa = 11.84IVDD172 pKa = 3.5SLLSPNSLAVLIAAVAGEE190 pKa = 4.43GSSVATDD197 pKa = 3.42ILRR200 pKa = 11.84LDD202 pKa = 3.35VSTRR206 pKa = 11.84KK207 pKa = 10.22AIVPIVSGPGLATAIVDD224 pKa = 3.99ALRR227 pKa = 11.84VLASNMVEE235 pKa = 3.92AGQGQLFSFAVTRR248 pKa = 11.84GLHH251 pKa = 4.48QALNVSSGTDD261 pKa = 3.31GQYY264 pKa = 11.57VMQSILRR271 pKa = 11.84SGRR274 pKa = 11.84FSPPLGGIHH283 pKa = 6.52TSLPVFSGLPALVSEE298 pKa = 4.53SRR300 pKa = 11.84RR301 pKa = 11.84DD302 pKa = 3.25VATFVDD308 pKa = 4.36ALALVSAAGAAPADD322 pKa = 3.62PCIVEE327 pKa = 4.51RR328 pKa = 11.84GNTFPTVLVSPGQISDD344 pKa = 4.3LDD346 pKa = 4.16SSSSMPGAFPSTPQLLGQLKK366 pKa = 9.23EE367 pKa = 3.91ALPAFFSNYY376 pKa = 9.25LRR378 pKa = 11.84ILSTVFAAGEE388 pKa = 4.14SDD390 pKa = 5.53SVALDD395 pKa = 3.72CACAFASLLEE405 pKa = 4.53DD406 pKa = 4.15LPTPLSAPTVLPFFWVEE423 pKa = 3.93PTSLLPASVFSTTAEE438 pKa = 4.11SAGFGALATPGSPVTRR454 pKa = 11.84PAWGAIEE461 pKa = 4.86HH462 pKa = 6.22LVSPSSAVSTATVSWTSARR481 pKa = 11.84QNPFLWHH488 pKa = 5.69FKK490 pKa = 10.27GMRR493 pKa = 11.84SDD495 pKa = 3.57PLAAVIVDD503 pKa = 3.69QFDD506 pKa = 3.72PTDD509 pKa = 3.48LAFAGPGGAEE519 pKa = 3.84TTLQKK524 pKa = 9.89WGRR527 pKa = 11.84QAPLTDD533 pKa = 3.68YY534 pKa = 10.92LWRR537 pKa = 11.84DD538 pKa = 3.42SASCLPAPGEE548 pKa = 4.0LLNLSGAIGLRR559 pKa = 11.84FRR561 pKa = 11.84HH562 pKa = 5.95FVIDD566 pKa = 5.25DD567 pKa = 3.81NDD569 pKa = 3.55DD570 pKa = 3.58CAFTRR575 pKa = 11.84IPMFHH580 pKa = 7.4EE581 pKa = 4.6IGTGSVTIAVSRR593 pKa = 11.84PCGLAPDD600 pKa = 5.05GPPARR605 pKa = 11.84LEE607 pKa = 4.0RR608 pKa = 11.84ATVVSPALGYY618 pKa = 10.33LRR620 pKa = 11.84QRR622 pKa = 11.84GANLRR627 pKa = 11.84AFGRR631 pKa = 11.84PDD633 pKa = 3.8DD634 pKa = 4.36LLAPRR639 pKa = 11.84RR640 pKa = 11.84LGPASLTVAPKK651 pKa = 10.54APPAPVSAPVSSFAFSGPKK670 pKa = 9.08TSSRR674 pKa = 11.84DD675 pKa = 3.62PLSGEE680 pKa = 4.14LPTPLVPVQHH690 pKa = 6.38YY691 pKa = 9.74KK692 pKa = 11.03AEE694 pKa = 4.42TGPKK698 pKa = 9.27TGGLGAAGGGGGTVKK713 pKa = 10.43PPTAVPPGLTEE724 pKa = 4.37VPTAPDD730 pKa = 3.61AGPTPEE736 pKa = 4.16PHH738 pKa = 6.31GAAPSLHH745 pKa = 6.22EE746 pKa = 4.14
MM1 pKa = 7.18AQIGAPNFLSSVLGDD16 pKa = 3.38QRR18 pKa = 11.84GGVLNSDD25 pKa = 3.67SVFRR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 9.22RR32 pKa = 11.84AAITTSTIVRR42 pKa = 11.84GVQDD46 pKa = 3.41TRR48 pKa = 11.84VGFLVFAVGRR58 pKa = 11.84VFSSFSAALVRR69 pKa = 11.84PKK71 pKa = 10.85SSVPAIDD78 pKa = 3.32ATYY81 pKa = 8.75PCPATRR87 pKa = 11.84SEE89 pKa = 4.4EE90 pKa = 4.11FVGLAKK96 pKa = 10.32KK97 pKa = 10.47YY98 pKa = 10.76SNFSSTFTHH107 pKa = 6.79ANFAGVVEE115 pKa = 4.22RR116 pKa = 11.84LSRR119 pKa = 11.84ALAVLSTLPDD129 pKa = 3.42SDD131 pKa = 3.95TAALTSLDD139 pKa = 3.22IAGGVMPTVYY149 pKa = 10.55SIATFDD155 pKa = 3.86SPVNALARR163 pKa = 11.84VVFIPRR169 pKa = 11.84IVDD172 pKa = 3.5SLLSPNSLAVLIAAVAGEE190 pKa = 4.43GSSVATDD197 pKa = 3.42ILRR200 pKa = 11.84LDD202 pKa = 3.35VSTRR206 pKa = 11.84KK207 pKa = 10.22AIVPIVSGPGLATAIVDD224 pKa = 3.99ALRR227 pKa = 11.84VLASNMVEE235 pKa = 3.92AGQGQLFSFAVTRR248 pKa = 11.84GLHH251 pKa = 4.48QALNVSSGTDD261 pKa = 3.31GQYY264 pKa = 11.57VMQSILRR271 pKa = 11.84SGRR274 pKa = 11.84FSPPLGGIHH283 pKa = 6.52TSLPVFSGLPALVSEE298 pKa = 4.53SRR300 pKa = 11.84RR301 pKa = 11.84DD302 pKa = 3.25VATFVDD308 pKa = 4.36ALALVSAAGAAPADD322 pKa = 3.62PCIVEE327 pKa = 4.51RR328 pKa = 11.84GNTFPTVLVSPGQISDD344 pKa = 4.3LDD346 pKa = 4.16SSSSMPGAFPSTPQLLGQLKK366 pKa = 9.23EE367 pKa = 3.91ALPAFFSNYY376 pKa = 9.25LRR378 pKa = 11.84ILSTVFAAGEE388 pKa = 4.14SDD390 pKa = 5.53SVALDD395 pKa = 3.72CACAFASLLEE405 pKa = 4.53DD406 pKa = 4.15LPTPLSAPTVLPFFWVEE423 pKa = 3.93PTSLLPASVFSTTAEE438 pKa = 4.11SAGFGALATPGSPVTRR454 pKa = 11.84PAWGAIEE461 pKa = 4.86HH462 pKa = 6.22LVSPSSAVSTATVSWTSARR481 pKa = 11.84QNPFLWHH488 pKa = 5.69FKK490 pKa = 10.27GMRR493 pKa = 11.84SDD495 pKa = 3.57PLAAVIVDD503 pKa = 3.69QFDD506 pKa = 3.72PTDD509 pKa = 3.48LAFAGPGGAEE519 pKa = 3.84TTLQKK524 pKa = 9.89WGRR527 pKa = 11.84QAPLTDD533 pKa = 3.68YY534 pKa = 10.92LWRR537 pKa = 11.84DD538 pKa = 3.42SASCLPAPGEE548 pKa = 4.0LLNLSGAIGLRR559 pKa = 11.84FRR561 pKa = 11.84HH562 pKa = 5.95FVIDD566 pKa = 5.25DD567 pKa = 3.81NDD569 pKa = 3.55DD570 pKa = 3.58CAFTRR575 pKa = 11.84IPMFHH580 pKa = 7.4EE581 pKa = 4.6IGTGSVTIAVSRR593 pKa = 11.84PCGLAPDD600 pKa = 5.05GPPARR605 pKa = 11.84LEE607 pKa = 4.0RR608 pKa = 11.84ATVVSPALGYY618 pKa = 10.33LRR620 pKa = 11.84QRR622 pKa = 11.84GANLRR627 pKa = 11.84AFGRR631 pKa = 11.84PDD633 pKa = 3.8DD634 pKa = 4.36LLAPRR639 pKa = 11.84RR640 pKa = 11.84LGPASLTVAPKK651 pKa = 10.54APPAPVSAPVSSFAFSGPKK670 pKa = 9.08TSSRR674 pKa = 11.84DD675 pKa = 3.62PLSGEE680 pKa = 4.14LPTPLVPVQHH690 pKa = 6.38YY691 pKa = 9.74KK692 pKa = 11.03AEE694 pKa = 4.42TGPKK698 pKa = 9.27TGGLGAAGGGGGTVKK713 pKa = 10.43PPTAVPPGLTEE724 pKa = 4.37VPTAPDD730 pKa = 3.61AGPTPEE736 pKa = 4.16PHH738 pKa = 6.31GAAPSLHH745 pKa = 6.22EE746 pKa = 4.14
Molecular weight: 77.22 kDa
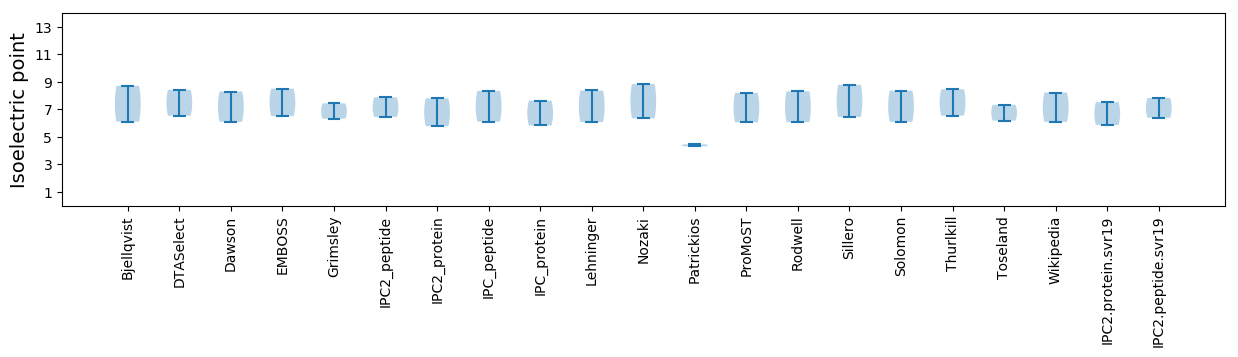
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q60GI3|Q60GI3_9VIRU Putative coat protein OS=Magnaporthe oryzae virus 1 OX=271257 PE=4 SV=1
MM1 pKa = 7.69DD2 pKa = 5.05LEE4 pKa = 4.55ARR6 pKa = 11.84SKK8 pKa = 10.93SFGRR12 pKa = 11.84LGMSCSSIYY21 pKa = 10.65SKK23 pKa = 11.19YY24 pKa = 9.88RR25 pKa = 11.84DD26 pKa = 3.87HH27 pKa = 7.49LPVFSSMDD35 pKa = 3.56FLSGLTYY42 pKa = 9.28VTTNVGVLRR51 pKa = 11.84SHH53 pKa = 7.18HH54 pKa = 6.85PLLPTAVSLLCLDD67 pKa = 4.93FPIQFDD73 pKa = 3.88TSSSHH78 pKa = 6.92CLLLAQSAYY87 pKa = 7.27MTVDD91 pKa = 3.37EE92 pKa = 5.1LARR95 pKa = 11.84SYY97 pKa = 10.44PCKK100 pKa = 10.35FGGRR104 pKa = 11.84RR105 pKa = 11.84ILGRR109 pKa = 11.84LIHH112 pKa = 7.31DD113 pKa = 3.64STFRR117 pKa = 11.84EE118 pKa = 3.85ATFPVKK124 pKa = 10.31SHH126 pKa = 6.99PAAHH130 pKa = 6.09TKK132 pKa = 10.57AHH134 pKa = 6.54ISLGALLRR142 pKa = 11.84SWPSVAPRR150 pKa = 11.84SEE152 pKa = 3.23IDD154 pKa = 3.69YY155 pKa = 10.95ILARR159 pKa = 11.84CAGRR163 pKa = 11.84LTADD167 pKa = 3.16QAIALIVYY175 pKa = 8.56HH176 pKa = 6.41VSLRR180 pKa = 11.84PTFGLSSARR189 pKa = 11.84FAVAALMCPGDD200 pKa = 4.35AKK202 pKa = 11.07GLSNALKK209 pKa = 10.48ALGLNGVKK217 pKa = 10.17EE218 pKa = 4.16GAVLVEE224 pKa = 4.0AQTLQGRR231 pKa = 11.84GVAPIDD237 pKa = 3.38WGRR240 pKa = 11.84EE241 pKa = 3.73IPSRR245 pKa = 11.84CTDD248 pKa = 3.06AVHH251 pKa = 6.62EE252 pKa = 4.29NTVFIPEE259 pKa = 4.02ADD261 pKa = 3.39LRR263 pKa = 11.84AHH265 pKa = 5.77VRR267 pKa = 11.84HH268 pKa = 5.91FLSSEE273 pKa = 3.85LDD275 pKa = 3.17ADD277 pKa = 4.13TSLEE281 pKa = 4.21PLDD284 pKa = 4.31HH285 pKa = 5.7WWSRR289 pKa = 11.84RR290 pKa = 11.84WAWCVNGAHH299 pKa = 6.4TSAASRR305 pKa = 11.84ALGIDD310 pKa = 3.22HH311 pKa = 7.0RR312 pKa = 11.84HH313 pKa = 6.07AFPAHH318 pKa = 5.17SRR320 pKa = 11.84VYY322 pKa = 10.73RR323 pKa = 11.84RR324 pKa = 11.84MASEE328 pKa = 3.98ALEE331 pKa = 4.2SEE333 pKa = 4.87PVSKK337 pKa = 10.24WDD339 pKa = 3.06GTTFVSASEE348 pKa = 4.02KK349 pKa = 10.7LEE351 pKa = 4.25HH352 pKa = 6.45GKK354 pKa = 7.98TRR356 pKa = 11.84AIFACDD362 pKa = 3.15TRR364 pKa = 11.84SYY366 pKa = 10.37FAWSWLLDD374 pKa = 3.46PVAANWRR381 pKa = 11.84NSRR384 pKa = 11.84ILLDD388 pKa = 3.81PGRR391 pKa = 11.84GGTYY395 pKa = 9.99GSTRR399 pKa = 11.84RR400 pKa = 11.84IQNAQLTGGVNVMLDD415 pKa = 3.43YY416 pKa = 11.58DD417 pKa = 4.55DD418 pKa = 6.12FNSQHH423 pKa = 5.82STRR426 pKa = 11.84SMQIVTEE433 pKa = 4.19EE434 pKa = 3.98LCSYY438 pKa = 10.37IGAPQWYY445 pKa = 8.19TDD447 pKa = 3.79VLVKK451 pKa = 10.81SLDD454 pKa = 3.45SEE456 pKa = 5.17YY457 pKa = 9.72ITGHH461 pKa = 5.54GPPRR465 pKa = 11.84HH466 pKa = 5.19VAGTLMSGHH475 pKa = 7.16RR476 pKa = 11.84GTTFFNSILNGVYY489 pKa = 9.45IRR491 pKa = 11.84HH492 pKa = 6.27AFGAGAFDD500 pKa = 4.15SCVSMHH506 pKa = 6.58TGDD509 pKa = 3.88DD510 pKa = 3.25VYY512 pKa = 11.08MRR514 pKa = 11.84LRR516 pKa = 11.84TLRR519 pKa = 11.84DD520 pKa = 3.05ASTLLVSLKK529 pKa = 10.93DD530 pKa = 3.3LGCRR534 pKa = 11.84LNPTKK539 pKa = 10.55QSIGYY544 pKa = 8.49KK545 pKa = 8.88HH546 pKa = 6.75AEE548 pKa = 4.12FLRR551 pKa = 11.84VAITPTGSRR560 pKa = 11.84GYY562 pKa = 10.19AARR565 pKa = 11.84SIAALASGNWTDD577 pKa = 4.49SDD579 pKa = 4.15PMDD582 pKa = 4.2ARR584 pKa = 11.84DD585 pKa = 4.11GFTTTLANIRR595 pKa = 11.84TIQLRR600 pKa = 11.84SGSPDD605 pKa = 3.02LGRR608 pKa = 11.84LVGPAVKK615 pKa = 10.11YY616 pKa = 10.89ARR618 pKa = 11.84GLRR621 pKa = 11.84TRR623 pKa = 11.84TVIKK627 pKa = 10.07ILNGEE632 pKa = 3.99EE633 pKa = 4.49SYY635 pKa = 11.35NGSPVYY641 pKa = 10.07NVPGRR646 pKa = 11.84TVKK649 pKa = 10.24VHH651 pKa = 5.82TFAPKK656 pKa = 10.61DD657 pKa = 3.63PDD659 pKa = 3.57YY660 pKa = 9.72MLPEE664 pKa = 4.9IPVSSSWPLNSTYY677 pKa = 10.86DD678 pKa = 3.88YY679 pKa = 11.33LNSHH683 pKa = 5.92CTGLEE688 pKa = 3.73SRR690 pKa = 11.84LFEE693 pKa = 4.19IAQVSPVPLLAASSYY708 pKa = 11.33SKK710 pKa = 11.14GLAQSRR716 pKa = 11.84QVLTSADD723 pKa = 3.51LTIRR727 pKa = 11.84PLGTVSTLGVIDD739 pKa = 4.1VGSLSAAPPSPPVLAGHH756 pKa = 6.88PLARR760 pKa = 11.84LVAARR765 pKa = 11.84VSDD768 pKa = 3.89DD769 pKa = 3.8DD770 pKa = 4.15LRR772 pKa = 11.84EE773 pKa = 4.04ILDD776 pKa = 3.57QFAIPPPPADD786 pKa = 3.27MTVRR790 pKa = 11.84EE791 pKa = 4.2YY792 pKa = 11.44AFGYY796 pKa = 9.73EE797 pKa = 3.87PHH799 pKa = 6.65CVNVRR804 pKa = 11.84GFLPHH809 pKa = 7.27ADD811 pKa = 3.55ASSLSARR818 pKa = 11.84TRR820 pKa = 11.84AGTIFVPTPVNVV832 pKa = 3.25
MM1 pKa = 7.69DD2 pKa = 5.05LEE4 pKa = 4.55ARR6 pKa = 11.84SKK8 pKa = 10.93SFGRR12 pKa = 11.84LGMSCSSIYY21 pKa = 10.65SKK23 pKa = 11.19YY24 pKa = 9.88RR25 pKa = 11.84DD26 pKa = 3.87HH27 pKa = 7.49LPVFSSMDD35 pKa = 3.56FLSGLTYY42 pKa = 9.28VTTNVGVLRR51 pKa = 11.84SHH53 pKa = 7.18HH54 pKa = 6.85PLLPTAVSLLCLDD67 pKa = 4.93FPIQFDD73 pKa = 3.88TSSSHH78 pKa = 6.92CLLLAQSAYY87 pKa = 7.27MTVDD91 pKa = 3.37EE92 pKa = 5.1LARR95 pKa = 11.84SYY97 pKa = 10.44PCKK100 pKa = 10.35FGGRR104 pKa = 11.84RR105 pKa = 11.84ILGRR109 pKa = 11.84LIHH112 pKa = 7.31DD113 pKa = 3.64STFRR117 pKa = 11.84EE118 pKa = 3.85ATFPVKK124 pKa = 10.31SHH126 pKa = 6.99PAAHH130 pKa = 6.09TKK132 pKa = 10.57AHH134 pKa = 6.54ISLGALLRR142 pKa = 11.84SWPSVAPRR150 pKa = 11.84SEE152 pKa = 3.23IDD154 pKa = 3.69YY155 pKa = 10.95ILARR159 pKa = 11.84CAGRR163 pKa = 11.84LTADD167 pKa = 3.16QAIALIVYY175 pKa = 8.56HH176 pKa = 6.41VSLRR180 pKa = 11.84PTFGLSSARR189 pKa = 11.84FAVAALMCPGDD200 pKa = 4.35AKK202 pKa = 11.07GLSNALKK209 pKa = 10.48ALGLNGVKK217 pKa = 10.17EE218 pKa = 4.16GAVLVEE224 pKa = 4.0AQTLQGRR231 pKa = 11.84GVAPIDD237 pKa = 3.38WGRR240 pKa = 11.84EE241 pKa = 3.73IPSRR245 pKa = 11.84CTDD248 pKa = 3.06AVHH251 pKa = 6.62EE252 pKa = 4.29NTVFIPEE259 pKa = 4.02ADD261 pKa = 3.39LRR263 pKa = 11.84AHH265 pKa = 5.77VRR267 pKa = 11.84HH268 pKa = 5.91FLSSEE273 pKa = 3.85LDD275 pKa = 3.17ADD277 pKa = 4.13TSLEE281 pKa = 4.21PLDD284 pKa = 4.31HH285 pKa = 5.7WWSRR289 pKa = 11.84RR290 pKa = 11.84WAWCVNGAHH299 pKa = 6.4TSAASRR305 pKa = 11.84ALGIDD310 pKa = 3.22HH311 pKa = 7.0RR312 pKa = 11.84HH313 pKa = 6.07AFPAHH318 pKa = 5.17SRR320 pKa = 11.84VYY322 pKa = 10.73RR323 pKa = 11.84RR324 pKa = 11.84MASEE328 pKa = 3.98ALEE331 pKa = 4.2SEE333 pKa = 4.87PVSKK337 pKa = 10.24WDD339 pKa = 3.06GTTFVSASEE348 pKa = 4.02KK349 pKa = 10.7LEE351 pKa = 4.25HH352 pKa = 6.45GKK354 pKa = 7.98TRR356 pKa = 11.84AIFACDD362 pKa = 3.15TRR364 pKa = 11.84SYY366 pKa = 10.37FAWSWLLDD374 pKa = 3.46PVAANWRR381 pKa = 11.84NSRR384 pKa = 11.84ILLDD388 pKa = 3.81PGRR391 pKa = 11.84GGTYY395 pKa = 9.99GSTRR399 pKa = 11.84RR400 pKa = 11.84IQNAQLTGGVNVMLDD415 pKa = 3.43YY416 pKa = 11.58DD417 pKa = 4.55DD418 pKa = 6.12FNSQHH423 pKa = 5.82STRR426 pKa = 11.84SMQIVTEE433 pKa = 4.19EE434 pKa = 3.98LCSYY438 pKa = 10.37IGAPQWYY445 pKa = 8.19TDD447 pKa = 3.79VLVKK451 pKa = 10.81SLDD454 pKa = 3.45SEE456 pKa = 5.17YY457 pKa = 9.72ITGHH461 pKa = 5.54GPPRR465 pKa = 11.84HH466 pKa = 5.19VAGTLMSGHH475 pKa = 7.16RR476 pKa = 11.84GTTFFNSILNGVYY489 pKa = 9.45IRR491 pKa = 11.84HH492 pKa = 6.27AFGAGAFDD500 pKa = 4.15SCVSMHH506 pKa = 6.58TGDD509 pKa = 3.88DD510 pKa = 3.25VYY512 pKa = 11.08MRR514 pKa = 11.84LRR516 pKa = 11.84TLRR519 pKa = 11.84DD520 pKa = 3.05ASTLLVSLKK529 pKa = 10.93DD530 pKa = 3.3LGCRR534 pKa = 11.84LNPTKK539 pKa = 10.55QSIGYY544 pKa = 8.49KK545 pKa = 8.88HH546 pKa = 6.75AEE548 pKa = 4.12FLRR551 pKa = 11.84VAITPTGSRR560 pKa = 11.84GYY562 pKa = 10.19AARR565 pKa = 11.84SIAALASGNWTDD577 pKa = 4.49SDD579 pKa = 4.15PMDD582 pKa = 4.2ARR584 pKa = 11.84DD585 pKa = 4.11GFTTTLANIRR595 pKa = 11.84TIQLRR600 pKa = 11.84SGSPDD605 pKa = 3.02LGRR608 pKa = 11.84LVGPAVKK615 pKa = 10.11YY616 pKa = 10.89ARR618 pKa = 11.84GLRR621 pKa = 11.84TRR623 pKa = 11.84TVIKK627 pKa = 10.07ILNGEE632 pKa = 3.99EE633 pKa = 4.49SYY635 pKa = 11.35NGSPVYY641 pKa = 10.07NVPGRR646 pKa = 11.84TVKK649 pKa = 10.24VHH651 pKa = 5.82TFAPKK656 pKa = 10.61DD657 pKa = 3.63PDD659 pKa = 3.57YY660 pKa = 9.72MLPEE664 pKa = 4.9IPVSSSWPLNSTYY677 pKa = 10.86DD678 pKa = 3.88YY679 pKa = 11.33LNSHH683 pKa = 5.92CTGLEE688 pKa = 3.73SRR690 pKa = 11.84LFEE693 pKa = 4.19IAQVSPVPLLAASSYY708 pKa = 11.33SKK710 pKa = 11.14GLAQSRR716 pKa = 11.84QVLTSADD723 pKa = 3.51LTIRR727 pKa = 11.84PLGTVSTLGVIDD739 pKa = 4.1VGSLSAAPPSPPVLAGHH756 pKa = 6.88PLARR760 pKa = 11.84LVAARR765 pKa = 11.84VSDD768 pKa = 3.89DD769 pKa = 3.8DD770 pKa = 4.15LRR772 pKa = 11.84EE773 pKa = 4.04ILDD776 pKa = 3.57QFAIPPPPADD786 pKa = 3.27MTVRR790 pKa = 11.84EE791 pKa = 4.2YY792 pKa = 11.44AFGYY796 pKa = 9.73EE797 pKa = 3.87PHH799 pKa = 6.65CVNVRR804 pKa = 11.84GFLPHH809 pKa = 7.27ADD811 pKa = 3.55ASSLSARR818 pKa = 11.84TRR820 pKa = 11.84AGTIFVPTPVNVV832 pKa = 3.25
Molecular weight: 90.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1578 |
746 |
832 |
789.0 |
84.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.217 ± 1.029 | 1.331 ± 0.266 |
5.26 ± 0.294 | 3.169 ± 0.149 |
4.246 ± 0.575 | 7.858 ± 0.489 |
2.535 ± 0.81 | 3.802 ± 0.306 |
2.028 ± 0.284 | 10.266 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.331 ± 0.266 | 2.281 ± 0.274 |
7.668 ± 1.254 | 2.091 ± 0.127 |
6.591 ± 0.742 | 10.33 ± 0.176 |
6.781 ± 0.129 | 7.731 ± 0.666 |
1.204 ± 0.271 | 2.281 ± 0.729 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
