
Pigeon adenovirus 2a
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Aviadenovirus; unclassified Aviadenovirus
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
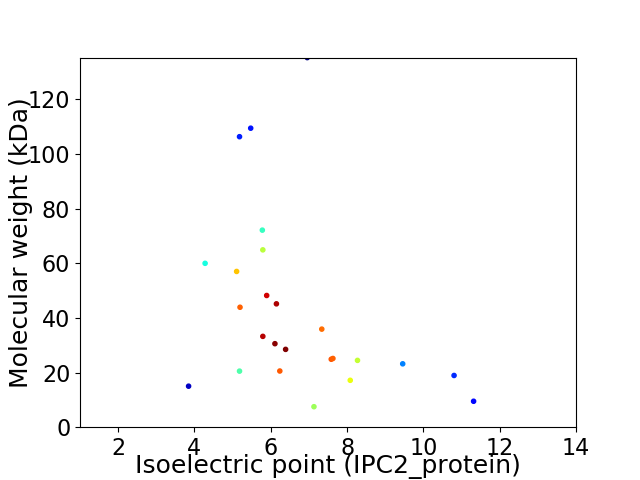
Virtual 2D-PAGE plot for 25 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L5LC42|A0A1L5LC42_9ADEN DBP OS=Pigeon adenovirus 2a OX=1924072 PE=4 SV=1
MM1 pKa = 7.65LKK3 pKa = 10.1RR4 pKa = 11.84RR5 pKa = 11.84NEE7 pKa = 3.83LSDD10 pKa = 3.14DD11 pKa = 4.19RR12 pKa = 11.84KK13 pKa = 10.18RR14 pKa = 11.84KK15 pKa = 9.36RR16 pKa = 11.84PRR18 pKa = 11.84AVGSSDD24 pKa = 4.68SINLNYY30 pKa = 9.41PFWYY34 pKa = 9.82SAEE37 pKa = 4.14EE38 pKa = 3.89NTEE41 pKa = 3.79IVPPFIDD48 pKa = 4.02PSGPLYY54 pKa = 11.08DD55 pKa = 4.43QDD57 pKa = 3.35GKK59 pKa = 11.68LNIRR63 pKa = 11.84LTSPIALISNGVGLGYY79 pKa = 10.57DD80 pKa = 3.24SSLEE84 pKa = 4.05NVNNRR89 pKa = 11.84LGVRR93 pKa = 11.84IDD95 pKa = 3.22SDD97 pKa = 3.74GALEE101 pKa = 4.04VTPDD105 pKa = 3.61GLNVRR110 pKa = 11.84VDD112 pKa = 3.86DD113 pKa = 4.13EE114 pKa = 4.92TVGINPSWEE123 pKa = 4.11LAVQFADD130 pKa = 3.62EE131 pKa = 4.65QPFSTSTAGIGLNVDD146 pKa = 3.52DD147 pKa = 4.99TLLISSSDD155 pKa = 3.56SDD157 pKa = 3.71PSHH160 pKa = 6.89YY161 pKa = 10.61EE162 pKa = 3.7LGVHH166 pKa = 6.93LNTEE170 pKa = 4.43GPITADD176 pKa = 3.27EE177 pKa = 4.35NGIDD181 pKa = 5.11LDD183 pKa = 4.42FNPHH187 pKa = 5.43TLTVQTNEE195 pKa = 3.79QNQGILSVFLKK206 pKa = 11.14ANGGLGATEE215 pKa = 4.03SGIGVNYY222 pKa = 10.22DD223 pKa = 3.59EE224 pKa = 5.45QSLHH228 pKa = 6.86LDD230 pKa = 3.38SGNDD234 pKa = 3.37LAVRR238 pKa = 11.84FAPEE242 pKa = 3.6QPFTVTPQGITLNIDD257 pKa = 3.28PSSLQVQNGTLSALTSGLGLDD278 pKa = 3.99IDD280 pKa = 4.54NNTLQVDD287 pKa = 4.1TVNNKK292 pKa = 6.99EE293 pKa = 4.27TLSVRR298 pKa = 11.84IAPDD302 pKa = 3.29NPVEE306 pKa = 4.46ADD308 pKa = 3.14SSGIKK313 pKa = 10.23LLYY316 pKa = 8.76NTGDD320 pKa = 3.49FFDD323 pKa = 4.64NGQSGLSAVTPISYY337 pKa = 10.42LSPYY341 pKa = 9.7CYY343 pKa = 10.64YY344 pKa = 10.63EE345 pKa = 5.86AGDD348 pKa = 4.19SSLTDD353 pKa = 3.16YY354 pKa = 11.34SSTVRR359 pKa = 11.84SSGNTNWPIAAYY371 pKa = 10.26VKK373 pKa = 10.21VANCSGVCNGVLNFVLNRR391 pKa = 11.84NNITFLGTTNDD402 pKa = 3.39SNNNYY407 pKa = 10.0VKK409 pKa = 10.57FCFVLNPGGNLDD421 pKa = 4.61GIHH424 pKa = 6.73SYY426 pKa = 8.73VHH428 pKa = 5.79NNIYY432 pKa = 10.29FPAGANTPTFMAPTKK447 pKa = 9.93TPPPNQTIPANWANWFVAKK466 pKa = 9.62WVGVRR471 pKa = 11.84VRR473 pKa = 11.84FYY475 pKa = 10.93ATDD478 pKa = 3.19SSGNQAIEE486 pKa = 4.66SYY488 pKa = 8.98VHH490 pKa = 6.43YY491 pKa = 10.79YY492 pKa = 8.77PALVNGSSGYY502 pKa = 9.52PVIFFVFEE510 pKa = 3.55IRR512 pKa = 11.84LPSGQNWYY520 pKa = 8.53TGSGLSTDD528 pKa = 3.95YY529 pKa = 11.1EE530 pKa = 4.32ILNSGPIPFQYY541 pKa = 10.88LATKK545 pKa = 10.08PSYY548 pKa = 9.65TAVSQQ553 pKa = 3.51
MM1 pKa = 7.65LKK3 pKa = 10.1RR4 pKa = 11.84RR5 pKa = 11.84NEE7 pKa = 3.83LSDD10 pKa = 3.14DD11 pKa = 4.19RR12 pKa = 11.84KK13 pKa = 10.18RR14 pKa = 11.84KK15 pKa = 9.36RR16 pKa = 11.84PRR18 pKa = 11.84AVGSSDD24 pKa = 4.68SINLNYY30 pKa = 9.41PFWYY34 pKa = 9.82SAEE37 pKa = 4.14EE38 pKa = 3.89NTEE41 pKa = 3.79IVPPFIDD48 pKa = 4.02PSGPLYY54 pKa = 11.08DD55 pKa = 4.43QDD57 pKa = 3.35GKK59 pKa = 11.68LNIRR63 pKa = 11.84LTSPIALISNGVGLGYY79 pKa = 10.57DD80 pKa = 3.24SSLEE84 pKa = 4.05NVNNRR89 pKa = 11.84LGVRR93 pKa = 11.84IDD95 pKa = 3.22SDD97 pKa = 3.74GALEE101 pKa = 4.04VTPDD105 pKa = 3.61GLNVRR110 pKa = 11.84VDD112 pKa = 3.86DD113 pKa = 4.13EE114 pKa = 4.92TVGINPSWEE123 pKa = 4.11LAVQFADD130 pKa = 3.62EE131 pKa = 4.65QPFSTSTAGIGLNVDD146 pKa = 3.52DD147 pKa = 4.99TLLISSSDD155 pKa = 3.56SDD157 pKa = 3.71PSHH160 pKa = 6.89YY161 pKa = 10.61EE162 pKa = 3.7LGVHH166 pKa = 6.93LNTEE170 pKa = 4.43GPITADD176 pKa = 3.27EE177 pKa = 4.35NGIDD181 pKa = 5.11LDD183 pKa = 4.42FNPHH187 pKa = 5.43TLTVQTNEE195 pKa = 3.79QNQGILSVFLKK206 pKa = 11.14ANGGLGATEE215 pKa = 4.03SGIGVNYY222 pKa = 10.22DD223 pKa = 3.59EE224 pKa = 5.45QSLHH228 pKa = 6.86LDD230 pKa = 3.38SGNDD234 pKa = 3.37LAVRR238 pKa = 11.84FAPEE242 pKa = 3.6QPFTVTPQGITLNIDD257 pKa = 3.28PSSLQVQNGTLSALTSGLGLDD278 pKa = 3.99IDD280 pKa = 4.54NNTLQVDD287 pKa = 4.1TVNNKK292 pKa = 6.99EE293 pKa = 4.27TLSVRR298 pKa = 11.84IAPDD302 pKa = 3.29NPVEE306 pKa = 4.46ADD308 pKa = 3.14SSGIKK313 pKa = 10.23LLYY316 pKa = 8.76NTGDD320 pKa = 3.49FFDD323 pKa = 4.64NGQSGLSAVTPISYY337 pKa = 10.42LSPYY341 pKa = 9.7CYY343 pKa = 10.64YY344 pKa = 10.63EE345 pKa = 5.86AGDD348 pKa = 4.19SSLTDD353 pKa = 3.16YY354 pKa = 11.34SSTVRR359 pKa = 11.84SSGNTNWPIAAYY371 pKa = 10.26VKK373 pKa = 10.21VANCSGVCNGVLNFVLNRR391 pKa = 11.84NNITFLGTTNDD402 pKa = 3.39SNNNYY407 pKa = 10.0VKK409 pKa = 10.57FCFVLNPGGNLDD421 pKa = 4.61GIHH424 pKa = 6.73SYY426 pKa = 8.73VHH428 pKa = 5.79NNIYY432 pKa = 10.29FPAGANTPTFMAPTKK447 pKa = 9.93TPPPNQTIPANWANWFVAKK466 pKa = 9.62WVGVRR471 pKa = 11.84VRR473 pKa = 11.84FYY475 pKa = 10.93ATDD478 pKa = 3.19SSGNQAIEE486 pKa = 4.66SYY488 pKa = 8.98VHH490 pKa = 6.43YY491 pKa = 10.79YY492 pKa = 8.77PALVNGSSGYY502 pKa = 9.52PVIFFVFEE510 pKa = 3.55IRR512 pKa = 11.84LPSGQNWYY520 pKa = 8.53TGSGLSTDD528 pKa = 3.95YY529 pKa = 11.1EE530 pKa = 4.32ILNSGPIPFQYY541 pKa = 10.88LATKK545 pKa = 10.08PSYY548 pKa = 9.65TAVSQQ553 pKa = 3.51
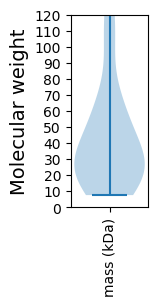
Molecular weight: 59.95 kDa
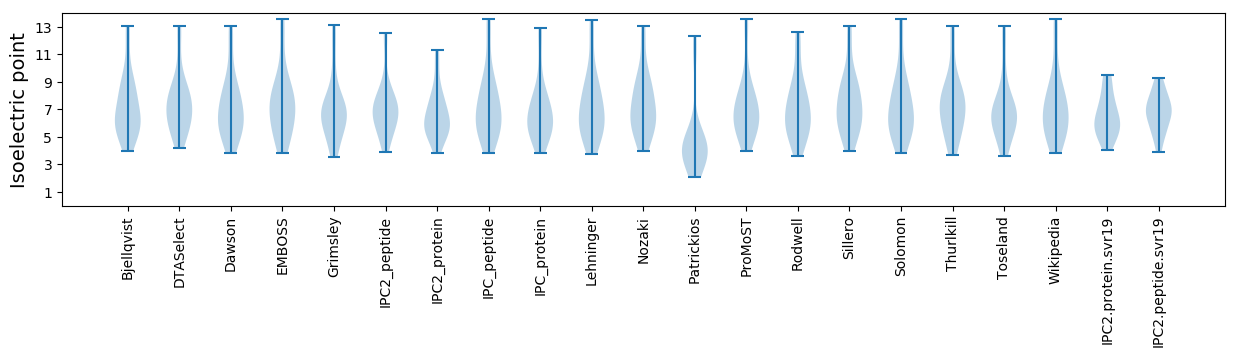
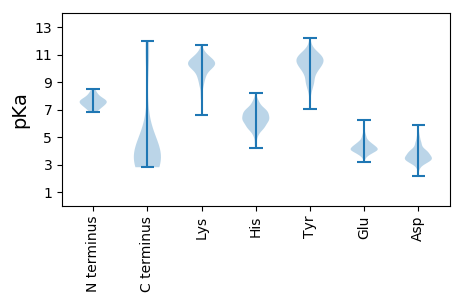
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L5LC33|A0A1L5LC33_9ADEN DNA polymerase OS=Pigeon adenovirus 2a OX=1924072 PE=3 SV=1
MM1 pKa = 7.74SILISPSDD9 pKa = 3.46NRR11 pKa = 11.84GWGMRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84TRR20 pKa = 11.84SSMRR24 pKa = 11.84GVGVRR29 pKa = 11.84RR30 pKa = 11.84PLTLRR35 pKa = 11.84RR36 pKa = 11.84LLGLGTTTSRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84NRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84AVRR56 pKa = 11.84GGASRR61 pKa = 11.84RR62 pKa = 11.84TSTSTRR68 pKa = 11.84IIAVRR73 pKa = 11.84TRR75 pKa = 11.84TSRR78 pKa = 11.84GRR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 3.26
MM1 pKa = 7.74SILISPSDD9 pKa = 3.46NRR11 pKa = 11.84GWGMRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84TRR20 pKa = 11.84SSMRR24 pKa = 11.84GVGVRR29 pKa = 11.84RR30 pKa = 11.84PLTLRR35 pKa = 11.84RR36 pKa = 11.84LLGLGTTTSRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84NRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84AVRR56 pKa = 11.84GGASRR61 pKa = 11.84RR62 pKa = 11.84TSTSTRR68 pKa = 11.84IIAVRR73 pKa = 11.84TRR75 pKa = 11.84TSRR78 pKa = 11.84GRR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 3.26
Molecular weight: 9.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9513 |
64 |
1176 |
380.5 |
43.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.728 ± 0.355 | 1.472 ± 0.247 |
5.603 ± 0.288 | 5.74 ± 0.528 |
4.383 ± 0.28 | 5.939 ± 0.422 |
2.439 ± 0.267 | 4.468 ± 0.205 |
3.784 ± 0.36 | 9.524 ± 0.484 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.47 ± 0.226 | 5.172 ± 0.566 |
6.097 ± 0.314 | 4.005 ± 0.243 |
7.043 ± 0.614 | 7.358 ± 0.351 |
6.234 ± 0.303 | 6.118 ± 0.224 |
1.314 ± 0.115 | 4.11 ± 0.353 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
