
Dendrolimus punctatus cypovirus 22
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Cypovirus; unclassified Cypovirus
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
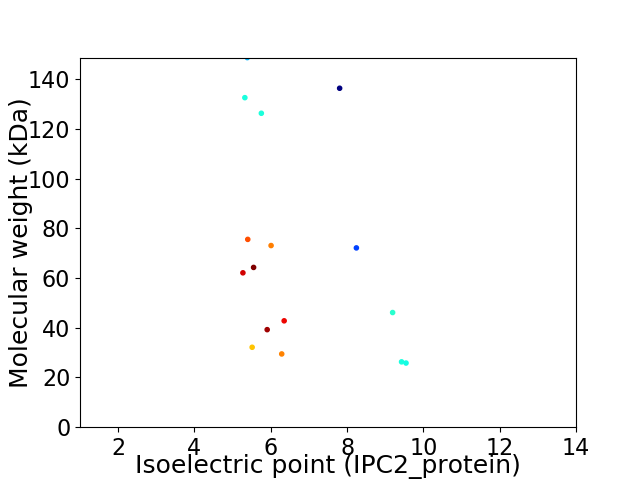
Virtual 2D-PAGE plot for 16 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
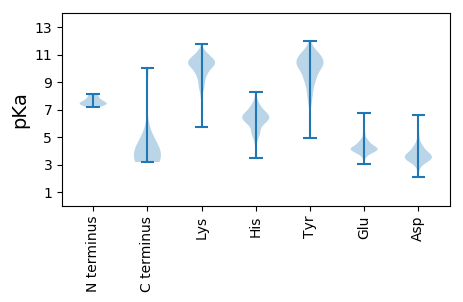
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A7E6B9|A0A0A7E6B9_9REOV Uncharacterized protein OS=Dendrolimus punctatus cypovirus 22 OX=1577776 PE=4 SV=1
MM1 pKa = 8.15EE2 pKa = 4.61YY3 pKa = 9.76PQSGFFAAEE12 pKa = 4.01TPNDD16 pKa = 3.2NRR18 pKa = 11.84TITLLQVRR26 pKa = 11.84GQQPRR31 pKa = 11.84NIPITDD37 pKa = 3.43ILDD40 pKa = 4.31HH41 pKa = 7.46IEE43 pKa = 4.51GPNTDD48 pKa = 4.3EE49 pKa = 4.75LFEE52 pKa = 4.39IQDD55 pKa = 3.55VTEE58 pKa = 4.19YY59 pKa = 10.95EE60 pKa = 4.24PDD62 pKa = 3.47HH63 pKa = 6.01TTHH66 pKa = 6.5ATNTQVFNINVLLRR80 pKa = 11.84SVFLSEE86 pKa = 4.12MTNVSQIPHH95 pKa = 6.85RR96 pKa = 11.84FGIFNVMSAEE106 pKa = 4.15DD107 pKa = 3.86ATCVVQKK114 pKa = 10.84DD115 pKa = 3.62GTYY118 pKa = 9.01YY119 pKa = 11.21ASGKK123 pKa = 7.85TRR125 pKa = 11.84DD126 pKa = 3.83ADD128 pKa = 4.32SIPHH132 pKa = 5.1VYY134 pKa = 9.61IRR136 pKa = 11.84NFALCMLKK144 pKa = 10.11VAPYY148 pKa = 10.68LRR150 pKa = 11.84GDD152 pKa = 4.51RR153 pKa = 11.84INVFAQLTHH162 pKa = 6.56FMQTVRR168 pKa = 11.84RR169 pKa = 11.84NRR171 pKa = 11.84SKK173 pKa = 10.25FWLRR177 pKa = 11.84FNLSDD182 pKa = 5.15LNPLHH187 pKa = 7.12LSNQLSKK194 pKa = 11.29FNLFLLLLDD203 pKa = 4.32LANYY207 pKa = 8.56EE208 pKa = 4.23HH209 pKa = 6.57FQVNQSAMLLGSINACLVCLFAAGISDD236 pKa = 3.4AVEE239 pKa = 3.63RR240 pKa = 11.84TTIDD244 pKa = 3.2EE245 pKa = 4.24VKK247 pKa = 10.28YY248 pKa = 10.24LQFKK252 pKa = 10.11IPSPSEE258 pKa = 3.59NYY260 pKa = 9.08VRR262 pKa = 11.84KK263 pKa = 10.12DD264 pKa = 3.04LLQPFFHH271 pKa = 6.37SQRR274 pKa = 11.84AIFVGFLINAFIQPYY289 pKa = 9.01MIVAFPLEE297 pKa = 4.08VRR299 pKa = 11.84NQLFPSNRR307 pKa = 11.84VPVVRR312 pKa = 11.84EE313 pKa = 3.98DD314 pKa = 4.03EE315 pKa = 4.34PVSDD319 pKa = 4.41HH320 pKa = 8.04DD321 pKa = 4.88SGEE324 pKa = 3.82ASNFYY329 pKa = 10.91DD330 pKa = 5.06LYY332 pKa = 11.44VSLNYY337 pKa = 10.46GAFPSDD343 pKa = 2.83GGIYY347 pKa = 10.05KK348 pKa = 10.32YY349 pKa = 10.73LSNSDD354 pKa = 3.5DD355 pKa = 3.91VIYY358 pKa = 11.06LDD360 pKa = 5.15VFDD363 pKa = 5.03TMPFQQRR370 pKa = 11.84YY371 pKa = 4.96QKK373 pKa = 10.95RR374 pKa = 11.84NIRR377 pKa = 11.84GLFDD381 pKa = 3.6DD382 pKa = 4.64TLNTNLNVIRR392 pKa = 11.84VVHH395 pKa = 6.33HH396 pKa = 6.69EE397 pKa = 4.85DD398 pKa = 3.44YY399 pKa = 11.42DD400 pKa = 4.22SVLLLDD406 pKa = 4.12ILQPFNEE413 pKa = 4.28SLSRR417 pKa = 11.84CLLTRR422 pKa = 11.84YY423 pKa = 10.01IIDD426 pKa = 3.19RR427 pKa = 11.84RR428 pKa = 11.84MIAVIASSTPCGLSDD443 pKa = 3.76GQLIFNAHH451 pKa = 4.54QLEE454 pKa = 4.45VFEE457 pKa = 6.01LITEE461 pKa = 4.64RR462 pKa = 11.84YY463 pKa = 7.68MNDD466 pKa = 3.62VIRR469 pKa = 11.84HH470 pKa = 5.39IPDD473 pKa = 3.94GYY475 pKa = 11.45GLAEE479 pKa = 4.18DD480 pKa = 4.14RR481 pKa = 11.84ARR483 pKa = 11.84VLHH486 pKa = 6.83EE487 pKa = 3.95ILALQASYY495 pKa = 11.06INEE498 pKa = 3.54IKK500 pKa = 8.11MTYY503 pKa = 10.39KK504 pKa = 10.7DD505 pKa = 3.48FSEE508 pKa = 4.75AYY510 pKa = 8.7KK511 pKa = 10.7HH512 pKa = 5.01KK513 pKa = 9.39TRR515 pKa = 11.84SNVSLEE521 pKa = 4.06KK522 pKa = 10.13RR523 pKa = 11.84WNEE526 pKa = 3.22AGIRR530 pKa = 11.84EE531 pKa = 4.32FVTCGLMLL539 pKa = 4.57
MM1 pKa = 8.15EE2 pKa = 4.61YY3 pKa = 9.76PQSGFFAAEE12 pKa = 4.01TPNDD16 pKa = 3.2NRR18 pKa = 11.84TITLLQVRR26 pKa = 11.84GQQPRR31 pKa = 11.84NIPITDD37 pKa = 3.43ILDD40 pKa = 4.31HH41 pKa = 7.46IEE43 pKa = 4.51GPNTDD48 pKa = 4.3EE49 pKa = 4.75LFEE52 pKa = 4.39IQDD55 pKa = 3.55VTEE58 pKa = 4.19YY59 pKa = 10.95EE60 pKa = 4.24PDD62 pKa = 3.47HH63 pKa = 6.01TTHH66 pKa = 6.5ATNTQVFNINVLLRR80 pKa = 11.84SVFLSEE86 pKa = 4.12MTNVSQIPHH95 pKa = 6.85RR96 pKa = 11.84FGIFNVMSAEE106 pKa = 4.15DD107 pKa = 3.86ATCVVQKK114 pKa = 10.84DD115 pKa = 3.62GTYY118 pKa = 9.01YY119 pKa = 11.21ASGKK123 pKa = 7.85TRR125 pKa = 11.84DD126 pKa = 3.83ADD128 pKa = 4.32SIPHH132 pKa = 5.1VYY134 pKa = 9.61IRR136 pKa = 11.84NFALCMLKK144 pKa = 10.11VAPYY148 pKa = 10.68LRR150 pKa = 11.84GDD152 pKa = 4.51RR153 pKa = 11.84INVFAQLTHH162 pKa = 6.56FMQTVRR168 pKa = 11.84RR169 pKa = 11.84NRR171 pKa = 11.84SKK173 pKa = 10.25FWLRR177 pKa = 11.84FNLSDD182 pKa = 5.15LNPLHH187 pKa = 7.12LSNQLSKK194 pKa = 11.29FNLFLLLLDD203 pKa = 4.32LANYY207 pKa = 8.56EE208 pKa = 4.23HH209 pKa = 6.57FQVNQSAMLLGSINACLVCLFAAGISDD236 pKa = 3.4AVEE239 pKa = 3.63RR240 pKa = 11.84TTIDD244 pKa = 3.2EE245 pKa = 4.24VKK247 pKa = 10.28YY248 pKa = 10.24LQFKK252 pKa = 10.11IPSPSEE258 pKa = 3.59NYY260 pKa = 9.08VRR262 pKa = 11.84KK263 pKa = 10.12DD264 pKa = 3.04LLQPFFHH271 pKa = 6.37SQRR274 pKa = 11.84AIFVGFLINAFIQPYY289 pKa = 9.01MIVAFPLEE297 pKa = 4.08VRR299 pKa = 11.84NQLFPSNRR307 pKa = 11.84VPVVRR312 pKa = 11.84EE313 pKa = 3.98DD314 pKa = 4.03EE315 pKa = 4.34PVSDD319 pKa = 4.41HH320 pKa = 8.04DD321 pKa = 4.88SGEE324 pKa = 3.82ASNFYY329 pKa = 10.91DD330 pKa = 5.06LYY332 pKa = 11.44VSLNYY337 pKa = 10.46GAFPSDD343 pKa = 2.83GGIYY347 pKa = 10.05KK348 pKa = 10.32YY349 pKa = 10.73LSNSDD354 pKa = 3.5DD355 pKa = 3.91VIYY358 pKa = 11.06LDD360 pKa = 5.15VFDD363 pKa = 5.03TMPFQQRR370 pKa = 11.84YY371 pKa = 4.96QKK373 pKa = 10.95RR374 pKa = 11.84NIRR377 pKa = 11.84GLFDD381 pKa = 3.6DD382 pKa = 4.64TLNTNLNVIRR392 pKa = 11.84VVHH395 pKa = 6.33HH396 pKa = 6.69EE397 pKa = 4.85DD398 pKa = 3.44YY399 pKa = 11.42DD400 pKa = 4.22SVLLLDD406 pKa = 4.12ILQPFNEE413 pKa = 4.28SLSRR417 pKa = 11.84CLLTRR422 pKa = 11.84YY423 pKa = 10.01IIDD426 pKa = 3.19RR427 pKa = 11.84RR428 pKa = 11.84MIAVIASSTPCGLSDD443 pKa = 3.76GQLIFNAHH451 pKa = 4.54QLEE454 pKa = 4.45VFEE457 pKa = 6.01LITEE461 pKa = 4.64RR462 pKa = 11.84YY463 pKa = 7.68MNDD466 pKa = 3.62VIRR469 pKa = 11.84HH470 pKa = 5.39IPDD473 pKa = 3.94GYY475 pKa = 11.45GLAEE479 pKa = 4.18DD480 pKa = 4.14RR481 pKa = 11.84ARR483 pKa = 11.84VLHH486 pKa = 6.83EE487 pKa = 3.95ILALQASYY495 pKa = 11.06INEE498 pKa = 3.54IKK500 pKa = 8.11MTYY503 pKa = 10.39KK504 pKa = 10.7DD505 pKa = 3.48FSEE508 pKa = 4.75AYY510 pKa = 8.7KK511 pKa = 10.7HH512 pKa = 5.01KK513 pKa = 9.39TRR515 pKa = 11.84SNVSLEE521 pKa = 4.06KK522 pKa = 10.13RR523 pKa = 11.84WNEE526 pKa = 3.22AGIRR530 pKa = 11.84EE531 pKa = 4.32FVTCGLMLL539 pKa = 4.57
Molecular weight: 62.09 kDa
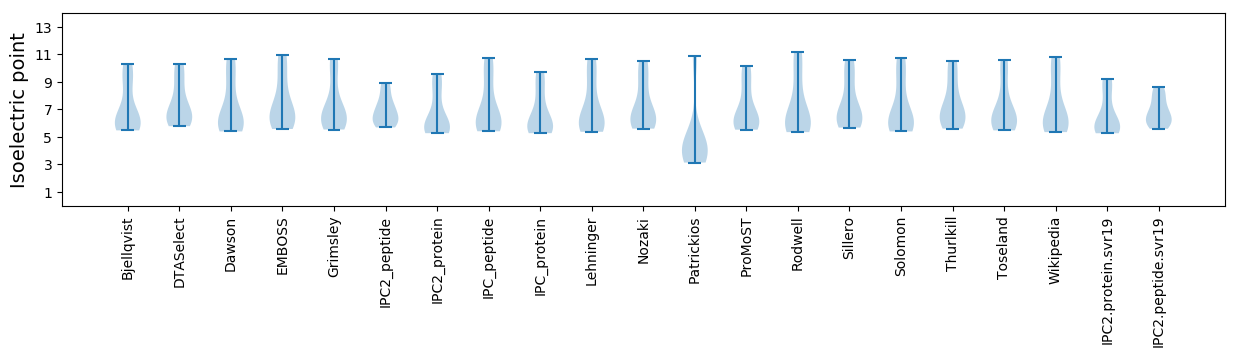
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A7E7V5|A0A0A7E7V5_9REOV Uncharacterized protein OS=Dendrolimus punctatus cypovirus 22 OX=1577776 PE=4 SV=1
MM1 pKa = 7.47KK2 pKa = 9.79KK3 pKa = 10.12KK4 pKa = 10.71SPASPLEE11 pKa = 4.16TLQSTMLDD19 pKa = 3.58CAKK22 pKa = 10.69LFDD25 pKa = 4.02RR26 pKa = 11.84TSKK29 pKa = 10.41FSKK32 pKa = 10.12AQRR35 pKa = 11.84LLAASLKK42 pKa = 10.34LIAEE46 pKa = 4.66IMPDD50 pKa = 2.77IPDD53 pKa = 3.22RR54 pKa = 11.84HH55 pKa = 5.84FPAIRR60 pKa = 11.84KK61 pKa = 5.72YY62 pKa = 9.19TKK64 pKa = 9.2ATKK67 pKa = 10.36NLATTAIKK75 pKa = 10.48NHH77 pKa = 6.31SLTQAKK83 pKa = 10.07AKK85 pKa = 10.28LIRR88 pKa = 11.84EE89 pKa = 3.85EE90 pKa = 4.35RR91 pKa = 11.84YY92 pKa = 9.9NDD94 pKa = 3.57RR95 pKa = 11.84VCIPNARR102 pKa = 11.84QVAATGDD109 pKa = 3.69YY110 pKa = 10.32QRR112 pKa = 11.84TSVVVRR118 pKa = 11.84STLRR122 pKa = 11.84ATLNDD127 pKa = 3.55VNDD130 pKa = 4.15FTANIEE136 pKa = 4.14YY137 pKa = 9.75HH138 pKa = 6.98RR139 pKa = 11.84KK140 pKa = 8.55RR141 pKa = 11.84QTVPEE146 pKa = 4.13LQRR149 pKa = 11.84EE150 pKa = 4.31LRR152 pKa = 11.84RR153 pKa = 11.84MNEE156 pKa = 3.86TLAEE160 pKa = 4.17LPALVQQGYY169 pKa = 8.96NRR171 pKa = 11.84YY172 pKa = 9.12CDD174 pKa = 3.99EE175 pKa = 4.71LRR177 pKa = 11.84HH178 pKa = 4.7PTIWKK183 pKa = 8.11QKK185 pKa = 8.93KK186 pKa = 8.0RR187 pKa = 11.84QKK189 pKa = 9.23EE190 pKa = 4.09TLTKK194 pKa = 10.39LEE196 pKa = 3.82KK197 pKa = 10.33SEE199 pKa = 4.43RR200 pKa = 11.84DD201 pKa = 3.2RR202 pKa = 11.84KK203 pKa = 10.82AKK205 pKa = 10.35FQVKK209 pKa = 9.6GGAGMWQRR217 pKa = 11.84LRR219 pKa = 11.84QQKK222 pKa = 10.04
MM1 pKa = 7.47KK2 pKa = 9.79KK3 pKa = 10.12KK4 pKa = 10.71SPASPLEE11 pKa = 4.16TLQSTMLDD19 pKa = 3.58CAKK22 pKa = 10.69LFDD25 pKa = 4.02RR26 pKa = 11.84TSKK29 pKa = 10.41FSKK32 pKa = 10.12AQRR35 pKa = 11.84LLAASLKK42 pKa = 10.34LIAEE46 pKa = 4.66IMPDD50 pKa = 2.77IPDD53 pKa = 3.22RR54 pKa = 11.84HH55 pKa = 5.84FPAIRR60 pKa = 11.84KK61 pKa = 5.72YY62 pKa = 9.19TKK64 pKa = 9.2ATKK67 pKa = 10.36NLATTAIKK75 pKa = 10.48NHH77 pKa = 6.31SLTQAKK83 pKa = 10.07AKK85 pKa = 10.28LIRR88 pKa = 11.84EE89 pKa = 3.85EE90 pKa = 4.35RR91 pKa = 11.84YY92 pKa = 9.9NDD94 pKa = 3.57RR95 pKa = 11.84VCIPNARR102 pKa = 11.84QVAATGDD109 pKa = 3.69YY110 pKa = 10.32QRR112 pKa = 11.84TSVVVRR118 pKa = 11.84STLRR122 pKa = 11.84ATLNDD127 pKa = 3.55VNDD130 pKa = 4.15FTANIEE136 pKa = 4.14YY137 pKa = 9.75HH138 pKa = 6.98RR139 pKa = 11.84KK140 pKa = 8.55RR141 pKa = 11.84QTVPEE146 pKa = 4.13LQRR149 pKa = 11.84EE150 pKa = 4.31LRR152 pKa = 11.84RR153 pKa = 11.84MNEE156 pKa = 3.86TLAEE160 pKa = 4.17LPALVQQGYY169 pKa = 8.96NRR171 pKa = 11.84YY172 pKa = 9.12CDD174 pKa = 3.99EE175 pKa = 4.71LRR177 pKa = 11.84HH178 pKa = 4.7PTIWKK183 pKa = 8.11QKK185 pKa = 8.93KK186 pKa = 8.0RR187 pKa = 11.84QKK189 pKa = 9.23EE190 pKa = 4.09TLTKK194 pKa = 10.39LEE196 pKa = 3.82KK197 pKa = 10.33SEE199 pKa = 4.43RR200 pKa = 11.84DD201 pKa = 3.2RR202 pKa = 11.84KK203 pKa = 10.82AKK205 pKa = 10.35FQVKK209 pKa = 9.6GGAGMWQRR217 pKa = 11.84LRR219 pKa = 11.84QQKK222 pKa = 10.04
Molecular weight: 25.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9936 |
222 |
1312 |
621.0 |
70.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.958 ± 0.173 | 1.329 ± 0.204 |
5.847 ± 0.211 | 5.304 ± 0.18 |
4.136 ± 0.258 | 4.64 ± 0.187 |
3.09 ± 0.183 | 7.377 ± 0.318 |
4.469 ± 0.471 | 9.541 ± 0.285 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.526 ± 0.162 | 5.435 ± 0.201 |
4.68 ± 0.281 | 4.147 ± 0.184 |
6.079 ± 0.329 | 6.924 ± 0.294 |
7.146 ± 0.362 | 6.159 ± 0.2 |
0.896 ± 0.069 | 4.318 ± 0.248 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
