
Cynara cardunculus var. scolymus (Globe artichoke) (Cynara scolymus)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; campanulids; Asterales; Asteraceae;
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
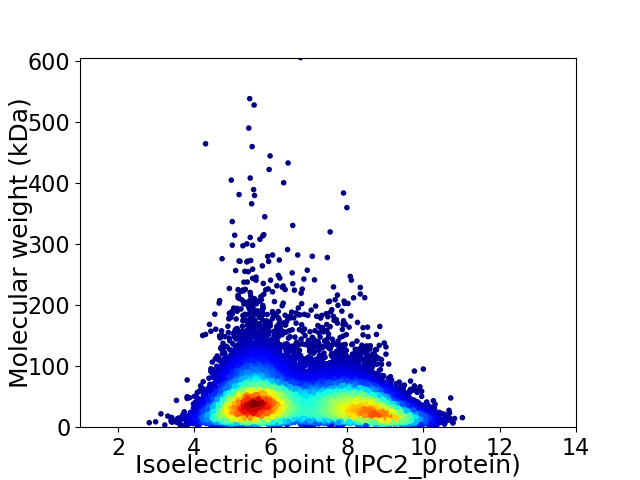
Virtual 2D-PAGE plot for 26434 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A103Y6C1|A0A103Y6C1_CYNCS Chlorophyll a-b binding protein chloroplastic OS=Cynara cardunculus var. scolymus OX=59895 GN=Ccrd_018365 PE=3 SV=1
MM1 pKa = 7.84ADD3 pKa = 3.33MSNIQALSLQPNTPQHH19 pKa = 5.26QVLEE23 pKa = 4.33EE24 pKa = 4.1EE25 pKa = 5.0EE26 pKa = 4.1EE27 pKa = 4.55DD28 pKa = 3.64EE29 pKa = 4.99TISSSQQTLDD39 pKa = 3.79NSITNCSSLVEE50 pKa = 4.22EE51 pKa = 5.06EE52 pKa = 4.57EE53 pKa = 5.24EE54 pKa = 4.87EE55 pKa = 5.71DD56 pKa = 5.8DD57 pKa = 6.63DD58 pKa = 7.0DD59 pKa = 6.54DD60 pKa = 5.33LVNVDD65 pKa = 4.39EE66 pKa = 5.49GDD68 pKa = 3.73VLPYY72 pKa = 10.07TFNFDD77 pKa = 3.47PFSPLNSSFFVPSHH91 pKa = 5.88HH92 pKa = 6.68HH93 pKa = 5.05HH94 pKa = 7.14HH95 pKa = 6.38NPQVNSNSCSCCSCEE110 pKa = 4.51SGSEE114 pKa = 4.19SEE116 pKa = 5.09SEE118 pKa = 4.07SDD120 pKa = 4.19CPDD123 pKa = 3.01STVYY127 pKa = 10.42PDD129 pKa = 3.62CLYY132 pKa = 11.21GHH134 pKa = 6.27EE135 pKa = 4.41EE136 pKa = 4.94EE137 pKa = 4.42YY138 pKa = 11.31DD139 pKa = 3.45QMDD142 pKa = 5.33FITDD146 pKa = 3.97LFDD149 pKa = 3.62SRR151 pKa = 11.84GEE153 pKa = 3.93IEE155 pKa = 4.9HH156 pKa = 6.89VSGTDD161 pKa = 3.06EE162 pKa = 4.94GISVSEE168 pKa = 5.32AINDD172 pKa = 3.73HH173 pKa = 7.35DD174 pKa = 5.73DD175 pKa = 4.32DD176 pKa = 4.46FVSSNIEE183 pKa = 3.72EE184 pKa = 4.52LGLGFEE190 pKa = 4.67GGGLRR195 pKa = 11.84VVGIEE200 pKa = 4.15SEE202 pKa = 4.33SDD204 pKa = 3.22SGTDD208 pKa = 3.79EE209 pKa = 4.64PGQVNDD215 pKa = 4.03QNDD218 pKa = 3.42EE219 pKa = 4.39RR220 pKa = 11.84IQITDD225 pKa = 3.33MNHH228 pKa = 3.45EE229 pKa = 4.76HH230 pKa = 6.75FWDD233 pKa = 4.55CPTFDD238 pKa = 3.57TARR241 pKa = 11.84VDD243 pKa = 4.47DD244 pKa = 4.58RR245 pKa = 11.84DD246 pKa = 3.95GLSSVIDD253 pKa = 4.33RR254 pKa = 11.84IEE256 pKa = 4.97DD257 pKa = 3.24ISVSSDD263 pKa = 2.46ISSEE267 pKa = 3.71GDD269 pKa = 3.43SAVGVLEE276 pKa = 3.99WEE278 pKa = 4.0ILLAVNNLEE287 pKa = 3.95RR288 pKa = 11.84DD289 pKa = 3.73LEE291 pKa = 4.37FEE293 pKa = 4.52TNGDD297 pKa = 3.38EE298 pKa = 4.45GFVYY302 pKa = 9.61TAEE305 pKa = 4.25FDD307 pKa = 3.69TLIGQFVEE315 pKa = 4.62TVRR318 pKa = 11.84ALRR321 pKa = 11.84GSPPAAKK328 pKa = 10.14SIIDD332 pKa = 3.69NLPSVVLKK340 pKa = 10.04TNNSKK345 pKa = 11.09DD346 pKa = 3.55GDD348 pKa = 4.1HH349 pKa = 7.48DD350 pKa = 3.91PVHH353 pKa = 6.33NQDD356 pKa = 3.84HH357 pKa = 6.73DD358 pKa = 4.05HH359 pKa = 6.79VICAVCKK366 pKa = 10.68DD367 pKa = 4.23EE368 pKa = 5.75ISMEE372 pKa = 4.2EE373 pKa = 4.56KK374 pKa = 9.6VTQLPCRR381 pKa = 11.84HH382 pKa = 6.75HH383 pKa = 5.75YY384 pKa = 10.7HH385 pKa = 7.05GDD387 pKa = 3.92CIVPWLSIRR396 pKa = 11.84NTCPVCRR403 pKa = 11.84FEE405 pKa = 6.22LPTDD409 pKa = 3.66DD410 pKa = 4.67VDD412 pKa = 4.25YY413 pKa = 10.85EE414 pKa = 4.17RR415 pKa = 11.84SKK417 pKa = 11.5NRR419 pKa = 11.84VNDD422 pKa = 3.38NEE424 pKa = 4.37LVVGYY429 pKa = 10.31GSEE432 pKa = 4.16LSPP435 pKa = 4.8
MM1 pKa = 7.84ADD3 pKa = 3.33MSNIQALSLQPNTPQHH19 pKa = 5.26QVLEE23 pKa = 4.33EE24 pKa = 4.1EE25 pKa = 5.0EE26 pKa = 4.1EE27 pKa = 4.55DD28 pKa = 3.64EE29 pKa = 4.99TISSSQQTLDD39 pKa = 3.79NSITNCSSLVEE50 pKa = 4.22EE51 pKa = 5.06EE52 pKa = 4.57EE53 pKa = 5.24EE54 pKa = 4.87EE55 pKa = 5.71DD56 pKa = 5.8DD57 pKa = 6.63DD58 pKa = 7.0DD59 pKa = 6.54DD60 pKa = 5.33LVNVDD65 pKa = 4.39EE66 pKa = 5.49GDD68 pKa = 3.73VLPYY72 pKa = 10.07TFNFDD77 pKa = 3.47PFSPLNSSFFVPSHH91 pKa = 5.88HH92 pKa = 6.68HH93 pKa = 5.05HH94 pKa = 7.14HH95 pKa = 6.38NPQVNSNSCSCCSCEE110 pKa = 4.51SGSEE114 pKa = 4.19SEE116 pKa = 5.09SEE118 pKa = 4.07SDD120 pKa = 4.19CPDD123 pKa = 3.01STVYY127 pKa = 10.42PDD129 pKa = 3.62CLYY132 pKa = 11.21GHH134 pKa = 6.27EE135 pKa = 4.41EE136 pKa = 4.94EE137 pKa = 4.42YY138 pKa = 11.31DD139 pKa = 3.45QMDD142 pKa = 5.33FITDD146 pKa = 3.97LFDD149 pKa = 3.62SRR151 pKa = 11.84GEE153 pKa = 3.93IEE155 pKa = 4.9HH156 pKa = 6.89VSGTDD161 pKa = 3.06EE162 pKa = 4.94GISVSEE168 pKa = 5.32AINDD172 pKa = 3.73HH173 pKa = 7.35DD174 pKa = 5.73DD175 pKa = 4.32DD176 pKa = 4.46FVSSNIEE183 pKa = 3.72EE184 pKa = 4.52LGLGFEE190 pKa = 4.67GGGLRR195 pKa = 11.84VVGIEE200 pKa = 4.15SEE202 pKa = 4.33SDD204 pKa = 3.22SGTDD208 pKa = 3.79EE209 pKa = 4.64PGQVNDD215 pKa = 4.03QNDD218 pKa = 3.42EE219 pKa = 4.39RR220 pKa = 11.84IQITDD225 pKa = 3.33MNHH228 pKa = 3.45EE229 pKa = 4.76HH230 pKa = 6.75FWDD233 pKa = 4.55CPTFDD238 pKa = 3.57TARR241 pKa = 11.84VDD243 pKa = 4.47DD244 pKa = 4.58RR245 pKa = 11.84DD246 pKa = 3.95GLSSVIDD253 pKa = 4.33RR254 pKa = 11.84IEE256 pKa = 4.97DD257 pKa = 3.24ISVSSDD263 pKa = 2.46ISSEE267 pKa = 3.71GDD269 pKa = 3.43SAVGVLEE276 pKa = 3.99WEE278 pKa = 4.0ILLAVNNLEE287 pKa = 3.95RR288 pKa = 11.84DD289 pKa = 3.73LEE291 pKa = 4.37FEE293 pKa = 4.52TNGDD297 pKa = 3.38EE298 pKa = 4.45GFVYY302 pKa = 9.61TAEE305 pKa = 4.25FDD307 pKa = 3.69TLIGQFVEE315 pKa = 4.62TVRR318 pKa = 11.84ALRR321 pKa = 11.84GSPPAAKK328 pKa = 10.14SIIDD332 pKa = 3.69NLPSVVLKK340 pKa = 10.04TNNSKK345 pKa = 11.09DD346 pKa = 3.55GDD348 pKa = 4.1HH349 pKa = 7.48DD350 pKa = 3.91PVHH353 pKa = 6.33NQDD356 pKa = 3.84HH357 pKa = 6.73DD358 pKa = 4.05HH359 pKa = 6.79VICAVCKK366 pKa = 10.68DD367 pKa = 4.23EE368 pKa = 5.75ISMEE372 pKa = 4.2EE373 pKa = 4.56KK374 pKa = 9.6VTQLPCRR381 pKa = 11.84HH382 pKa = 6.75HH383 pKa = 5.75YY384 pKa = 10.7HH385 pKa = 7.05GDD387 pKa = 3.92CIVPWLSIRR396 pKa = 11.84NTCPVCRR403 pKa = 11.84FEE405 pKa = 6.22LPTDD409 pKa = 3.66DD410 pKa = 4.67VDD412 pKa = 4.25YY413 pKa = 10.85EE414 pKa = 4.17RR415 pKa = 11.84SKK417 pKa = 11.5NRR419 pKa = 11.84VNDD422 pKa = 3.38NEE424 pKa = 4.37LVVGYY429 pKa = 10.31GSEE432 pKa = 4.16LSPP435 pKa = 4.8
Molecular weight: 48.43 kDa
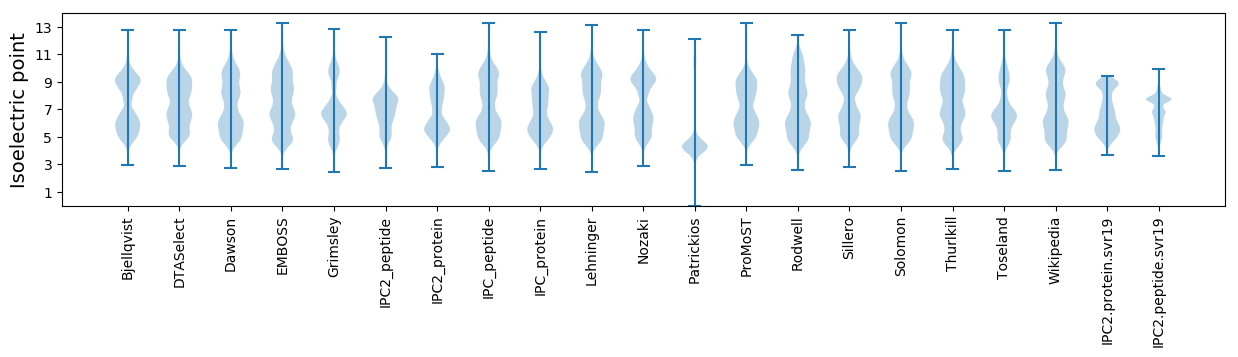
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A103Y954|A0A103Y954_CYNCS Protein kinase ATP binding site-containing protein OS=Cynara cardunculus var. scolymus OX=59895 GN=Ccrd_016883 PE=3 SV=1
MM1 pKa = 7.54LSLHH5 pKa = 6.65RR6 pKa = 11.84SSSVPIFKK14 pKa = 10.47LPSSSIVPLPSQSSSFHH31 pKa = 5.77RR32 pKa = 11.84HH33 pKa = 4.8PLFLFRR39 pKa = 11.84RR40 pKa = 11.84SSTRR44 pKa = 11.84IRR46 pKa = 11.84AMASNAVLAINLRR59 pKa = 11.84HH60 pKa = 5.79
MM1 pKa = 7.54LSLHH5 pKa = 6.65RR6 pKa = 11.84SSSVPIFKK14 pKa = 10.47LPSSSIVPLPSQSSSFHH31 pKa = 5.77RR32 pKa = 11.84HH33 pKa = 4.8PLFLFRR39 pKa = 11.84RR40 pKa = 11.84SSTRR44 pKa = 11.84IRR46 pKa = 11.84AMASNAVLAINLRR59 pKa = 11.84HH60 pKa = 5.79
Molecular weight: 6.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10709625 |
8 |
5364 |
405.1 |
45.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.461 ± 0.014 | 1.812 ± 0.008 |
5.39 ± 0.01 | 6.267 ± 0.017 |
4.278 ± 0.01 | 6.609 ± 0.014 |
2.455 ± 0.007 | 5.477 ± 0.011 |
6.074 ± 0.015 | 9.609 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.542 ± 0.007 | 4.538 ± 0.01 |
4.911 ± 0.017 | 3.57 ± 0.01 |
5.244 ± 0.01 | 8.951 ± 0.017 |
5.094 ± 0.009 | 6.587 ± 0.01 |
1.267 ± 0.006 | 2.831 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
