
Corynebacterium alimapuense
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Corynebacteriaceae; Corynebacterium
Average proteome isoelectric point is 5.73
Get precalculated fractions of proteins
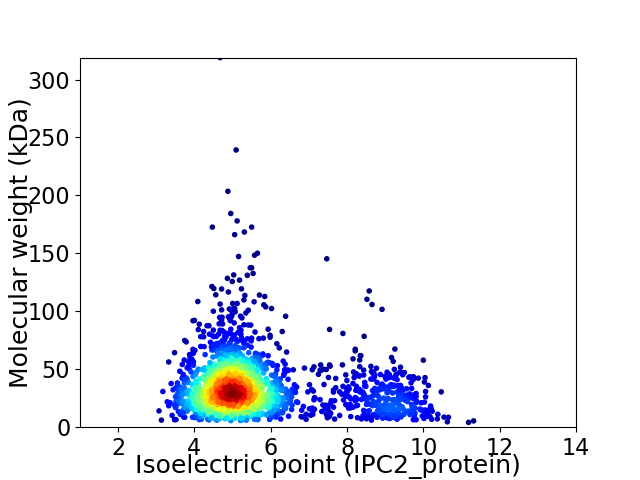
Virtual 2D-PAGE plot for 2040 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3M8K5W8|A0A3M8K5W8_9CORY SAF domain-containing protein OS=Corynebacterium alimapuense OX=1576874 GN=C5L39_08930 PE=4 SV=1
MM1 pKa = 7.93TYY3 pKa = 10.67RR4 pKa = 11.84KK5 pKa = 7.71TLGVLTATTLALSLVACSDD24 pKa = 3.95SEE26 pKa = 4.35GDD28 pKa = 3.85SSATGDD34 pKa = 3.6GSDD37 pKa = 3.28GGNYY41 pKa = 9.4VLANGSEE48 pKa = 4.18PQNPLIPANTNEE60 pKa = 4.07VGGGNIIDD68 pKa = 5.35LIFSGLVYY76 pKa = 10.61YY77 pKa = 10.69DD78 pKa = 3.84ADD80 pKa = 3.94GEE82 pKa = 4.38IHH84 pKa = 6.53NEE86 pKa = 3.76VAEE89 pKa = 4.41SIEE92 pKa = 4.07LEE94 pKa = 3.81GDD96 pKa = 2.74KK97 pKa = 10.45TYY99 pKa = 10.99RR100 pKa = 11.84VVLQDD105 pKa = 2.51GWTFNDD111 pKa = 4.23GSAVTADD118 pKa = 3.35SFVNAWNYY126 pKa = 10.43AVANSQLSSYY136 pKa = 10.16FFEE139 pKa = 5.83PILGYY144 pKa = 10.4EE145 pKa = 4.21EE146 pKa = 5.33GVEE149 pKa = 4.15SMEE152 pKa = 4.09GLTVIDD158 pKa = 4.27DD159 pKa = 3.79TTFTIEE165 pKa = 4.89LIEE168 pKa = 4.28PQADD172 pKa = 3.51FPTRR176 pKa = 11.84LGYY179 pKa = 10.24SAFYY183 pKa = 9.44PLPEE187 pKa = 4.0EE188 pKa = 5.25AFDD191 pKa = 4.62DD192 pKa = 3.78MAAFGEE198 pKa = 4.68TPNGNGPYY206 pKa = 10.2KK207 pKa = 10.44LSEE210 pKa = 3.99WNHH213 pKa = 5.25NQDD216 pKa = 3.02ATIVPSEE223 pKa = 4.24TYY225 pKa = 9.65EE226 pKa = 4.37GQRR229 pKa = 11.84TPQNDD234 pKa = 3.25GVKK237 pKa = 10.11FVFYY241 pKa = 10.21AQQDD245 pKa = 3.29AAYY248 pKa = 10.25NDD250 pKa = 4.23LLAGNLDD257 pKa = 3.9TLDD260 pKa = 5.46AIPDD264 pKa = 3.65SAFATFNEE272 pKa = 4.25EE273 pKa = 4.02LGDD276 pKa = 3.74RR277 pKa = 11.84AVNQPAAVFQSFTIPMEE294 pKa = 4.22AEE296 pKa = 3.96HH297 pKa = 6.8FSGEE301 pKa = 4.01EE302 pKa = 3.91GQLRR306 pKa = 11.84RR307 pKa = 11.84QALSLAINRR316 pKa = 11.84EE317 pKa = 4.34EE318 pKa = 3.6ITEE321 pKa = 4.45TIFQDD326 pKa = 3.14TRR328 pKa = 11.84TPASEE333 pKa = 3.89FTTPVLPGFDD343 pKa = 4.41DD344 pKa = 5.59EE345 pKa = 5.08IPGSEE350 pKa = 4.13VLDD353 pKa = 4.01YY354 pKa = 11.25DD355 pKa = 4.04PEE357 pKa = 4.34LANEE361 pKa = 4.63LWAQADD367 pKa = 4.67EE368 pKa = 4.19IDD370 pKa = 3.77PWSGEE375 pKa = 3.78FTLSYY380 pKa = 10.58NADD383 pKa = 3.71GGHH386 pKa = 6.65DD387 pKa = 3.11AWVNAVTNSIKK398 pKa = 9.68NTLGIEE404 pKa = 4.63AIGNPYY410 pKa = 10.09PDD412 pKa = 4.23FKK414 pKa = 11.17SLRR417 pKa = 11.84DD418 pKa = 3.62DD419 pKa = 3.41VTNRR423 pKa = 11.84TIEE426 pKa = 3.98GAFRR430 pKa = 11.84TGWQADD436 pKa = 3.95YY437 pKa = 10.05PSQGNFLGPLYY448 pKa = 10.0GTGAGSNDD456 pKa = 3.04GDD458 pKa = 3.81YY459 pKa = 11.48SNPEE463 pKa = 3.42FDD465 pKa = 5.1AKK467 pKa = 10.81LSEE470 pKa = 4.57ANSASSTEE478 pKa = 3.84EE479 pKa = 3.28SSRR482 pKa = 11.84IFNEE486 pKa = 3.78SQEE489 pKa = 4.26ILLSDD494 pKa = 5.0LPATPLWYY502 pKa = 10.78SNVAGGFSEE511 pKa = 5.09NVDD514 pKa = 2.97NVVFSWKK521 pKa = 10.01SVPVYY526 pKa = 10.54YY527 pKa = 10.45EE528 pKa = 3.38ITKK531 pKa = 10.19QQ532 pKa = 3.2
MM1 pKa = 7.93TYY3 pKa = 10.67RR4 pKa = 11.84KK5 pKa = 7.71TLGVLTATTLALSLVACSDD24 pKa = 3.95SEE26 pKa = 4.35GDD28 pKa = 3.85SSATGDD34 pKa = 3.6GSDD37 pKa = 3.28GGNYY41 pKa = 9.4VLANGSEE48 pKa = 4.18PQNPLIPANTNEE60 pKa = 4.07VGGGNIIDD68 pKa = 5.35LIFSGLVYY76 pKa = 10.61YY77 pKa = 10.69DD78 pKa = 3.84ADD80 pKa = 3.94GEE82 pKa = 4.38IHH84 pKa = 6.53NEE86 pKa = 3.76VAEE89 pKa = 4.41SIEE92 pKa = 4.07LEE94 pKa = 3.81GDD96 pKa = 2.74KK97 pKa = 10.45TYY99 pKa = 10.99RR100 pKa = 11.84VVLQDD105 pKa = 2.51GWTFNDD111 pKa = 4.23GSAVTADD118 pKa = 3.35SFVNAWNYY126 pKa = 10.43AVANSQLSSYY136 pKa = 10.16FFEE139 pKa = 5.83PILGYY144 pKa = 10.4EE145 pKa = 4.21EE146 pKa = 5.33GVEE149 pKa = 4.15SMEE152 pKa = 4.09GLTVIDD158 pKa = 4.27DD159 pKa = 3.79TTFTIEE165 pKa = 4.89LIEE168 pKa = 4.28PQADD172 pKa = 3.51FPTRR176 pKa = 11.84LGYY179 pKa = 10.24SAFYY183 pKa = 9.44PLPEE187 pKa = 4.0EE188 pKa = 5.25AFDD191 pKa = 4.62DD192 pKa = 3.78MAAFGEE198 pKa = 4.68TPNGNGPYY206 pKa = 10.2KK207 pKa = 10.44LSEE210 pKa = 3.99WNHH213 pKa = 5.25NQDD216 pKa = 3.02ATIVPSEE223 pKa = 4.24TYY225 pKa = 9.65EE226 pKa = 4.37GQRR229 pKa = 11.84TPQNDD234 pKa = 3.25GVKK237 pKa = 10.11FVFYY241 pKa = 10.21AQQDD245 pKa = 3.29AAYY248 pKa = 10.25NDD250 pKa = 4.23LLAGNLDD257 pKa = 3.9TLDD260 pKa = 5.46AIPDD264 pKa = 3.65SAFATFNEE272 pKa = 4.25EE273 pKa = 4.02LGDD276 pKa = 3.74RR277 pKa = 11.84AVNQPAAVFQSFTIPMEE294 pKa = 4.22AEE296 pKa = 3.96HH297 pKa = 6.8FSGEE301 pKa = 4.01EE302 pKa = 3.91GQLRR306 pKa = 11.84RR307 pKa = 11.84QALSLAINRR316 pKa = 11.84EE317 pKa = 4.34EE318 pKa = 3.6ITEE321 pKa = 4.45TIFQDD326 pKa = 3.14TRR328 pKa = 11.84TPASEE333 pKa = 3.89FTTPVLPGFDD343 pKa = 4.41DD344 pKa = 5.59EE345 pKa = 5.08IPGSEE350 pKa = 4.13VLDD353 pKa = 4.01YY354 pKa = 11.25DD355 pKa = 4.04PEE357 pKa = 4.34LANEE361 pKa = 4.63LWAQADD367 pKa = 4.67EE368 pKa = 4.19IDD370 pKa = 3.77PWSGEE375 pKa = 3.78FTLSYY380 pKa = 10.58NADD383 pKa = 3.71GGHH386 pKa = 6.65DD387 pKa = 3.11AWVNAVTNSIKK398 pKa = 9.68NTLGIEE404 pKa = 4.63AIGNPYY410 pKa = 10.09PDD412 pKa = 4.23FKK414 pKa = 11.17SLRR417 pKa = 11.84DD418 pKa = 3.62DD419 pKa = 3.41VTNRR423 pKa = 11.84TIEE426 pKa = 3.98GAFRR430 pKa = 11.84TGWQADD436 pKa = 3.95YY437 pKa = 10.05PSQGNFLGPLYY448 pKa = 10.0GTGAGSNDD456 pKa = 3.04GDD458 pKa = 3.81YY459 pKa = 11.48SNPEE463 pKa = 3.42FDD465 pKa = 5.1AKK467 pKa = 10.81LSEE470 pKa = 4.57ANSASSTEE478 pKa = 3.84EE479 pKa = 3.28SSRR482 pKa = 11.84IFNEE486 pKa = 3.78SQEE489 pKa = 4.26ILLSDD494 pKa = 5.0LPATPLWYY502 pKa = 10.78SNVAGGFSEE511 pKa = 5.09NVDD514 pKa = 2.97NVVFSWKK521 pKa = 10.01SVPVYY526 pKa = 10.54YY527 pKa = 10.45EE528 pKa = 3.38ITKK531 pKa = 10.19QQ532 pKa = 3.2
Molecular weight: 57.98 kDa
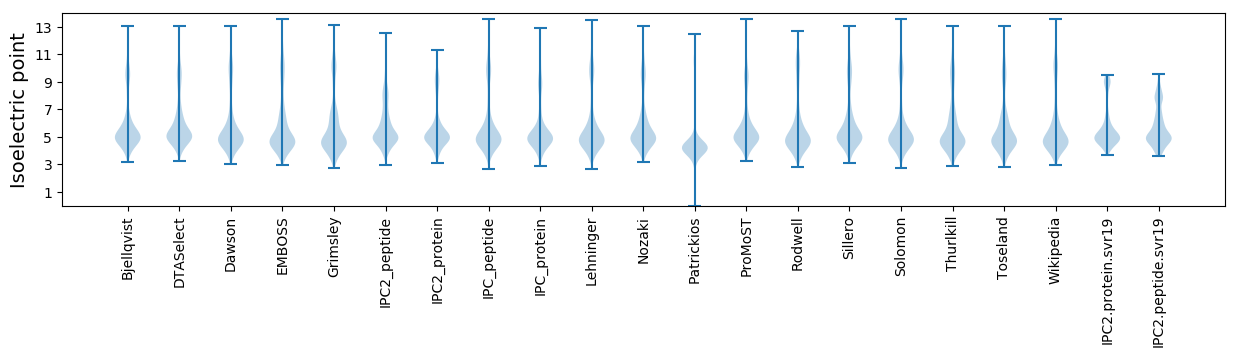
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3M8K5L2|A0A3M8K5L2_9CORY Non-specific serine/threonine protein kinase OS=Corynebacterium alimapuense OX=1576874 GN=pknB PE=4 SV=1
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.97GFRR22 pKa = 11.84TRR24 pKa = 11.84MSTRR28 pKa = 11.84AGRR31 pKa = 11.84AIVSARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.73GRR42 pKa = 11.84AKK44 pKa = 9.63LTAA47 pKa = 4.21
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.97GFRR22 pKa = 11.84TRR24 pKa = 11.84MSTRR28 pKa = 11.84AGRR31 pKa = 11.84AIVSARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.73GRR42 pKa = 11.84AKK44 pKa = 9.63LTAA47 pKa = 4.21
Molecular weight: 5.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
679389 |
33 |
3023 |
333.0 |
36.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.448 ± 0.07 | 0.653 ± 0.014 |
6.005 ± 0.047 | 6.369 ± 0.049 |
3.245 ± 0.029 | 8.373 ± 0.047 |
2.095 ± 0.026 | 5.368 ± 0.04 |
2.687 ± 0.041 | 10.155 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.185 ± 0.023 | 2.697 ± 0.031 |
4.976 ± 0.037 | 3.432 ± 0.03 |
6.164 ± 0.059 | 6.46 ± 0.043 |
6.012 ± 0.039 | 8.162 ± 0.047 |
1.412 ± 0.026 | 2.104 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
