
Filobacillus milosensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Filobacillus
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
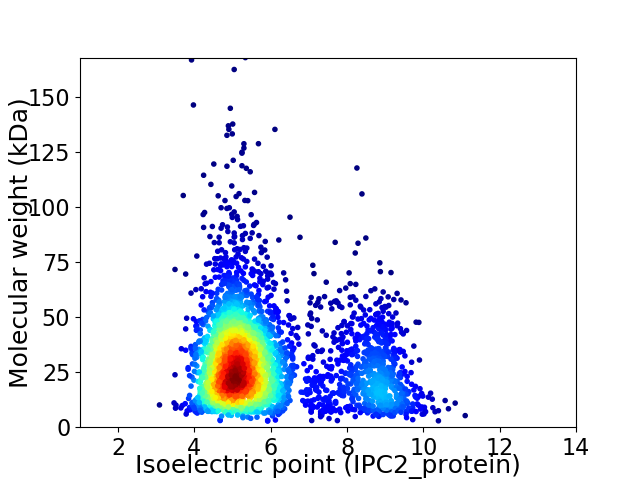
Virtual 2D-PAGE plot for 3330 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y8IF77|A0A4Y8IF77_9BACI Peptidase S8 OS=Filobacillus milosensis OX=94137 GN=E3U55_11320 PE=4 SV=1
MM1 pKa = 7.47AFIVGCVPDD10 pKa = 4.08EE11 pKa = 4.61APVEE15 pKa = 4.21DD16 pKa = 5.34DD17 pKa = 4.66EE18 pKa = 4.94NTEE21 pKa = 4.13EE22 pKa = 5.49DD23 pKa = 4.42NSDD26 pKa = 3.65SQNDD30 pKa = 3.89SEE32 pKa = 4.97NDD34 pKa = 3.33NSSDD38 pKa = 3.52EE39 pKa = 4.61GNLSGQLEE47 pKa = 3.97IQYY50 pKa = 10.25FVGGYY55 pKa = 9.85GDD57 pKa = 3.88SWWKK61 pKa = 10.22EE62 pKa = 3.68VIGDD66 pKa = 4.2FKK68 pKa = 11.37EE69 pKa = 4.18EE70 pKa = 4.14YY71 pKa = 9.8PNVEE75 pKa = 4.49VVEE78 pKa = 4.21HH79 pKa = 6.88AGPNINEE86 pKa = 4.23EE87 pKa = 4.01MRR89 pKa = 11.84SRR91 pKa = 11.84WVSGNPPDD99 pKa = 4.1VVYY102 pKa = 10.71IDD104 pKa = 4.16GAGSSEE110 pKa = 3.98TQMVEE115 pKa = 4.61DD116 pKa = 4.27GQLMNLSEE124 pKa = 4.08WAKK127 pKa = 10.54EE128 pKa = 3.74IEE130 pKa = 4.59LEE132 pKa = 5.13DD133 pKa = 3.84GTPLLDD139 pKa = 3.89NFIAEE144 pKa = 4.22PASYY148 pKa = 10.62DD149 pKa = 3.44GEE151 pKa = 4.79LYY153 pKa = 10.87SLPLVFDD160 pKa = 4.09TWGTWYY166 pKa = 10.48DD167 pKa = 3.68KK168 pKa = 11.41AWFEE172 pKa = 4.58DD173 pKa = 3.35GGYY176 pKa = 9.8EE177 pKa = 4.25VPKK180 pKa = 10.64DD181 pKa = 3.65FEE183 pKa = 4.97SFMNTMGDD191 pKa = 3.02IKK193 pKa = 10.65ASEE196 pKa = 5.13DD197 pKa = 3.2IPPFVTTGQYY207 pKa = 10.21PYY209 pKa = 10.81YY210 pKa = 9.85FLRR213 pKa = 11.84GMLYY217 pKa = 9.92PAFSAYY223 pKa = 10.3GGDD226 pKa = 3.63EE227 pKa = 4.06LLAGVITGEE236 pKa = 3.88EE237 pKa = 4.6GAWSSEE243 pKa = 3.81EE244 pKa = 3.98VLTVMQNVQRR254 pKa = 11.84MQEE257 pKa = 3.72EE258 pKa = 5.72GYY260 pKa = 10.14IDD262 pKa = 3.83PGFGAFNHH270 pKa = 6.2TQSQMNFLLHH280 pKa = 6.53EE281 pKa = 4.24NAFIPVGFWLPNEE294 pKa = 4.27MAGDD298 pKa = 3.83TPEE301 pKa = 3.79NFQYY305 pKa = 11.21GFIPSPMNEE314 pKa = 4.28AGEE317 pKa = 4.09PMAIVPDD324 pKa = 4.2LRR326 pKa = 11.84PLAIAEE332 pKa = 4.06EE333 pKa = 4.53AEE335 pKa = 4.03NPEE338 pKa = 3.89AAKK341 pKa = 10.89AFVDD345 pKa = 4.86FVFTRR350 pKa = 11.84EE351 pKa = 3.88YY352 pKa = 11.0AKK354 pKa = 10.7LFSEE358 pKa = 4.3HH359 pKa = 6.31TGAIMNISGVDD370 pKa = 3.47LSQNEE375 pKa = 4.3NVPEE379 pKa = 3.83YY380 pKa = 10.62LIEE383 pKa = 4.59ANAMINDD390 pKa = 4.1PEE392 pKa = 4.2QVQIYY397 pKa = 8.69HH398 pKa = 6.83KK399 pKa = 8.66PHH401 pKa = 6.75PMSADD406 pKa = 3.27LEE408 pKa = 4.72TPISNSLVSLMLGNITAEE426 pKa = 4.1KK427 pKa = 9.96FIEE430 pKa = 4.3EE431 pKa = 4.1AEE433 pKa = 4.38KK434 pKa = 10.69AAEE437 pKa = 4.18EE438 pKa = 4.14YY439 pKa = 10.84RR440 pKa = 11.84NNQQ443 pKa = 2.78
MM1 pKa = 7.47AFIVGCVPDD10 pKa = 4.08EE11 pKa = 4.61APVEE15 pKa = 4.21DD16 pKa = 5.34DD17 pKa = 4.66EE18 pKa = 4.94NTEE21 pKa = 4.13EE22 pKa = 5.49DD23 pKa = 4.42NSDD26 pKa = 3.65SQNDD30 pKa = 3.89SEE32 pKa = 4.97NDD34 pKa = 3.33NSSDD38 pKa = 3.52EE39 pKa = 4.61GNLSGQLEE47 pKa = 3.97IQYY50 pKa = 10.25FVGGYY55 pKa = 9.85GDD57 pKa = 3.88SWWKK61 pKa = 10.22EE62 pKa = 3.68VIGDD66 pKa = 4.2FKK68 pKa = 11.37EE69 pKa = 4.18EE70 pKa = 4.14YY71 pKa = 9.8PNVEE75 pKa = 4.49VVEE78 pKa = 4.21HH79 pKa = 6.88AGPNINEE86 pKa = 4.23EE87 pKa = 4.01MRR89 pKa = 11.84SRR91 pKa = 11.84WVSGNPPDD99 pKa = 4.1VVYY102 pKa = 10.71IDD104 pKa = 4.16GAGSSEE110 pKa = 3.98TQMVEE115 pKa = 4.61DD116 pKa = 4.27GQLMNLSEE124 pKa = 4.08WAKK127 pKa = 10.54EE128 pKa = 3.74IEE130 pKa = 4.59LEE132 pKa = 5.13DD133 pKa = 3.84GTPLLDD139 pKa = 3.89NFIAEE144 pKa = 4.22PASYY148 pKa = 10.62DD149 pKa = 3.44GEE151 pKa = 4.79LYY153 pKa = 10.87SLPLVFDD160 pKa = 4.09TWGTWYY166 pKa = 10.48DD167 pKa = 3.68KK168 pKa = 11.41AWFEE172 pKa = 4.58DD173 pKa = 3.35GGYY176 pKa = 9.8EE177 pKa = 4.25VPKK180 pKa = 10.64DD181 pKa = 3.65FEE183 pKa = 4.97SFMNTMGDD191 pKa = 3.02IKK193 pKa = 10.65ASEE196 pKa = 5.13DD197 pKa = 3.2IPPFVTTGQYY207 pKa = 10.21PYY209 pKa = 10.81YY210 pKa = 9.85FLRR213 pKa = 11.84GMLYY217 pKa = 9.92PAFSAYY223 pKa = 10.3GGDD226 pKa = 3.63EE227 pKa = 4.06LLAGVITGEE236 pKa = 3.88EE237 pKa = 4.6GAWSSEE243 pKa = 3.81EE244 pKa = 3.98VLTVMQNVQRR254 pKa = 11.84MQEE257 pKa = 3.72EE258 pKa = 5.72GYY260 pKa = 10.14IDD262 pKa = 3.83PGFGAFNHH270 pKa = 6.2TQSQMNFLLHH280 pKa = 6.53EE281 pKa = 4.24NAFIPVGFWLPNEE294 pKa = 4.27MAGDD298 pKa = 3.83TPEE301 pKa = 3.79NFQYY305 pKa = 11.21GFIPSPMNEE314 pKa = 4.28AGEE317 pKa = 4.09PMAIVPDD324 pKa = 4.2LRR326 pKa = 11.84PLAIAEE332 pKa = 4.06EE333 pKa = 4.53AEE335 pKa = 4.03NPEE338 pKa = 3.89AAKK341 pKa = 10.89AFVDD345 pKa = 4.86FVFTRR350 pKa = 11.84EE351 pKa = 3.88YY352 pKa = 11.0AKK354 pKa = 10.7LFSEE358 pKa = 4.3HH359 pKa = 6.31TGAIMNISGVDD370 pKa = 3.47LSQNEE375 pKa = 4.3NVPEE379 pKa = 3.83YY380 pKa = 10.62LIEE383 pKa = 4.59ANAMINDD390 pKa = 4.1PEE392 pKa = 4.2QVQIYY397 pKa = 8.69HH398 pKa = 6.83KK399 pKa = 8.66PHH401 pKa = 6.75PMSADD406 pKa = 3.27LEE408 pKa = 4.72TPISNSLVSLMLGNITAEE426 pKa = 4.1KK427 pKa = 9.96FIEE430 pKa = 4.3EE431 pKa = 4.1AEE433 pKa = 4.38KK434 pKa = 10.69AAEE437 pKa = 4.18EE438 pKa = 4.14YY439 pKa = 10.84RR440 pKa = 11.84NNQQ443 pKa = 2.78
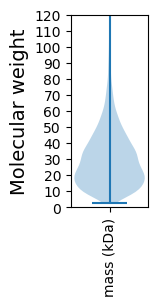
Molecular weight: 49.54 kDa
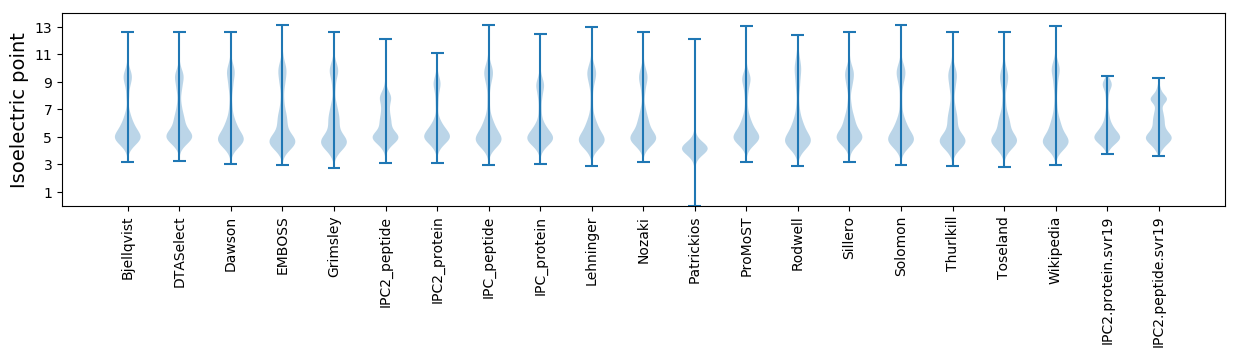
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y8IMI9|A0A4Y8IMI9_9BACI Uncharacterized protein OS=Filobacillus milosensis OX=94137 GN=E3U55_08950 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.35QPNNRR10 pKa = 11.84KK11 pKa = 9.28RR12 pKa = 11.84SKK14 pKa = 9.57VHH16 pKa = 5.93GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 10.31SKK25 pKa = 10.6NGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.36KK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.35QPNNRR10 pKa = 11.84KK11 pKa = 9.28RR12 pKa = 11.84SKK14 pKa = 9.57VHH16 pKa = 5.93GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 10.31SKK25 pKa = 10.6NGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.36KK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
921258 |
25 |
1532 |
276.7 |
31.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.11 ± 0.048 | 0.619 ± 0.013 |
5.692 ± 0.04 | 7.93 ± 0.053 |
4.608 ± 0.04 | 6.533 ± 0.038 |
2.159 ± 0.025 | 8.001 ± 0.039 |
6.796 ± 0.04 | 9.58 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.742 ± 0.02 | 4.975 ± 0.034 |
3.415 ± 0.024 | 4.04 ± 0.038 |
3.826 ± 0.032 | 5.987 ± 0.031 |
5.183 ± 0.025 | 7.028 ± 0.034 |
1.02 ± 0.017 | 3.756 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
