
Aeriscardovia aeriphila
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Bifidobacteriales; Bifidobacteriaceae; Aeriscardovia
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
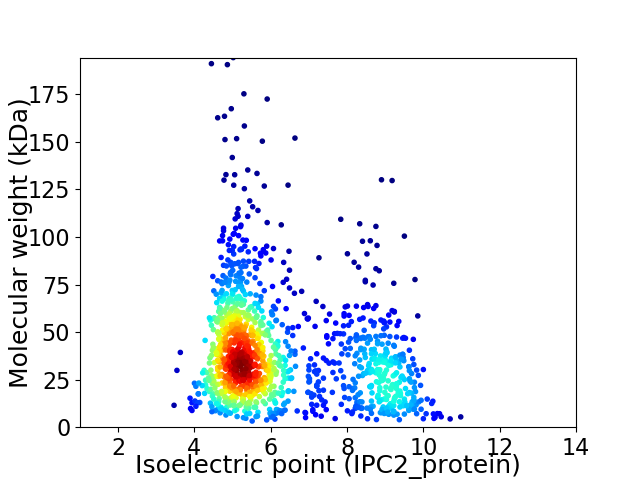
Virtual 2D-PAGE plot for 1287 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A261FC38|A0A261FC38_9BIFI DNA-binding protein OS=Aeriscardovia aeriphila OX=218139 GN=AEAE_0016 PE=4 SV=1
MM1 pKa = 7.61PLHH4 pKa = 5.42YY5 pKa = 9.71TSVADD10 pKa = 5.69AGIQPINLAQMRR22 pKa = 11.84QDD24 pKa = 3.02VHH26 pKa = 8.79LDD28 pKa = 3.5FTHH31 pKa = 7.03AACLLSVEE39 pKa = 4.42PEE41 pKa = 4.05VVEE44 pKa = 3.85RR45 pKa = 11.84WEE47 pKa = 4.25RR48 pKa = 11.84NLEE51 pKa = 4.03VLPVAVYY58 pKa = 9.99KK59 pKa = 10.74KK60 pKa = 10.63YY61 pKa = 10.94VATLEE66 pKa = 4.18KK67 pKa = 10.38AGRR70 pKa = 11.84IYY72 pKa = 10.68QQDD75 pKa = 3.72PDD77 pKa = 4.01NLIFTIDD84 pKa = 3.7LASLDD89 pKa = 4.2RR90 pKa = 11.84LDD92 pKa = 5.53HH93 pKa = 6.32PMTYY97 pKa = 9.03TACTEE102 pKa = 4.19EE103 pKa = 4.39YY104 pKa = 10.51VEE106 pKa = 4.34QVLALGVDD114 pKa = 4.49FPEE117 pKa = 5.22DD118 pKa = 3.53FWPYY122 pKa = 10.46EE123 pKa = 4.08FVPEE127 pKa = 3.97YY128 pKa = 10.33PIVLRR133 pKa = 11.84EE134 pKa = 3.9QIAYY138 pKa = 5.6EE139 pKa = 4.13TEE141 pKa = 4.54GIEE144 pKa = 4.2PYY146 pKa = 10.35PGFGEE151 pKa = 3.94AWEE154 pKa = 4.37TFQDD158 pKa = 3.81NLDD161 pKa = 3.79EE162 pKa = 4.42VDD164 pKa = 4.65SIVHH168 pKa = 6.43QILADD173 pKa = 3.42QGGKK177 pKa = 9.22VLYY180 pKa = 10.6DD181 pKa = 3.81DD182 pKa = 5.28DD183 pKa = 6.54DD184 pKa = 5.21YY185 pKa = 12.07YY186 pKa = 11.63GYY188 pKa = 10.86DD189 pKa = 4.07LPDD192 pKa = 3.45GVNFEE197 pKa = 4.36GDD199 pKa = 3.74SVALADD205 pKa = 5.05LARR208 pKa = 11.84EE209 pKa = 4.12NNTFDD214 pKa = 5.45DD215 pKa = 5.58DD216 pKa = 4.47STFDD220 pKa = 5.88DD221 pKa = 5.46DD222 pKa = 6.61DD223 pKa = 6.11FDD225 pKa = 6.61DD226 pKa = 5.83GSTFNDD232 pKa = 3.89GDD234 pKa = 3.94FEE236 pKa = 4.69DD237 pKa = 6.02ANDD240 pKa = 4.06ADD242 pKa = 4.97DD243 pKa = 5.82DD244 pKa = 4.49FSEE247 pKa = 5.22SYY249 pKa = 10.74TDD251 pKa = 4.6ANFADD256 pKa = 5.51FAARR260 pKa = 11.84LSQFSSGQEE269 pKa = 3.66DD270 pKa = 4.25GAPSLRR276 pKa = 11.84YY277 pKa = 10.12DD278 pKa = 3.04NGAIHH283 pKa = 6.73FEE285 pKa = 4.32VVDD288 pKa = 4.48LDD290 pKa = 3.78AGKK293 pKa = 10.18DD294 pKa = 3.27ADD296 pKa = 4.13TQTDD300 pKa = 3.32TGATRR305 pKa = 11.84SAALDD310 pKa = 3.67ADD312 pKa = 4.28TNADD316 pKa = 3.33AVTSANVNANAEE328 pKa = 4.26ANPIPITSARR338 pKa = 11.84IQRR341 pKa = 11.84GDD343 pKa = 3.21GSLSASHH350 pKa = 5.93STPPSEE356 pKa = 4.41
MM1 pKa = 7.61PLHH4 pKa = 5.42YY5 pKa = 9.71TSVADD10 pKa = 5.69AGIQPINLAQMRR22 pKa = 11.84QDD24 pKa = 3.02VHH26 pKa = 8.79LDD28 pKa = 3.5FTHH31 pKa = 7.03AACLLSVEE39 pKa = 4.42PEE41 pKa = 4.05VVEE44 pKa = 3.85RR45 pKa = 11.84WEE47 pKa = 4.25RR48 pKa = 11.84NLEE51 pKa = 4.03VLPVAVYY58 pKa = 9.99KK59 pKa = 10.74KK60 pKa = 10.63YY61 pKa = 10.94VATLEE66 pKa = 4.18KK67 pKa = 10.38AGRR70 pKa = 11.84IYY72 pKa = 10.68QQDD75 pKa = 3.72PDD77 pKa = 4.01NLIFTIDD84 pKa = 3.7LASLDD89 pKa = 4.2RR90 pKa = 11.84LDD92 pKa = 5.53HH93 pKa = 6.32PMTYY97 pKa = 9.03TACTEE102 pKa = 4.19EE103 pKa = 4.39YY104 pKa = 10.51VEE106 pKa = 4.34QVLALGVDD114 pKa = 4.49FPEE117 pKa = 5.22DD118 pKa = 3.53FWPYY122 pKa = 10.46EE123 pKa = 4.08FVPEE127 pKa = 3.97YY128 pKa = 10.33PIVLRR133 pKa = 11.84EE134 pKa = 3.9QIAYY138 pKa = 5.6EE139 pKa = 4.13TEE141 pKa = 4.54GIEE144 pKa = 4.2PYY146 pKa = 10.35PGFGEE151 pKa = 3.94AWEE154 pKa = 4.37TFQDD158 pKa = 3.81NLDD161 pKa = 3.79EE162 pKa = 4.42VDD164 pKa = 4.65SIVHH168 pKa = 6.43QILADD173 pKa = 3.42QGGKK177 pKa = 9.22VLYY180 pKa = 10.6DD181 pKa = 3.81DD182 pKa = 5.28DD183 pKa = 6.54DD184 pKa = 5.21YY185 pKa = 12.07YY186 pKa = 11.63GYY188 pKa = 10.86DD189 pKa = 4.07LPDD192 pKa = 3.45GVNFEE197 pKa = 4.36GDD199 pKa = 3.74SVALADD205 pKa = 5.05LARR208 pKa = 11.84EE209 pKa = 4.12NNTFDD214 pKa = 5.45DD215 pKa = 5.58DD216 pKa = 4.47STFDD220 pKa = 5.88DD221 pKa = 5.46DD222 pKa = 6.61DD223 pKa = 6.11FDD225 pKa = 6.61DD226 pKa = 5.83GSTFNDD232 pKa = 3.89GDD234 pKa = 3.94FEE236 pKa = 4.69DD237 pKa = 6.02ANDD240 pKa = 4.06ADD242 pKa = 4.97DD243 pKa = 5.82DD244 pKa = 4.49FSEE247 pKa = 5.22SYY249 pKa = 10.74TDD251 pKa = 4.6ANFADD256 pKa = 5.51FAARR260 pKa = 11.84LSQFSSGQEE269 pKa = 3.66DD270 pKa = 4.25GAPSLRR276 pKa = 11.84YY277 pKa = 10.12DD278 pKa = 3.04NGAIHH283 pKa = 6.73FEE285 pKa = 4.32VVDD288 pKa = 4.48LDD290 pKa = 3.78AGKK293 pKa = 10.18DD294 pKa = 3.27ADD296 pKa = 4.13TQTDD300 pKa = 3.32TGATRR305 pKa = 11.84SAALDD310 pKa = 3.67ADD312 pKa = 4.28TNADD316 pKa = 3.33AVTSANVNANAEE328 pKa = 4.26ANPIPITSARR338 pKa = 11.84IQRR341 pKa = 11.84GDD343 pKa = 3.21GSLSASHH350 pKa = 5.93STPPSEE356 pKa = 4.41
Molecular weight: 39.31 kDa
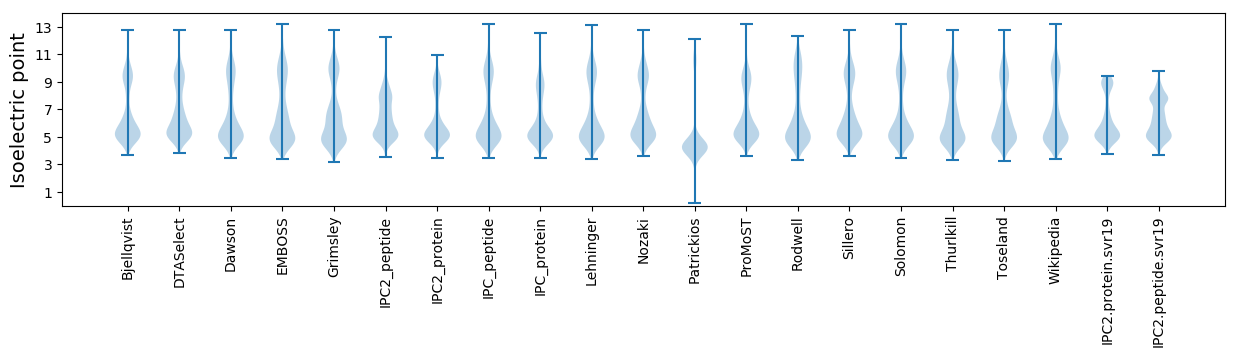
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A261F7X0|A0A261F7X0_9BIFI Ammonium transporter OS=Aeriscardovia aeriphila OX=218139 GN=AEAE_1215 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 4.8MKK15 pKa = 9.45HH16 pKa = 5.74GFRR19 pKa = 11.84EE20 pKa = 4.06RR21 pKa = 11.84MRR23 pKa = 11.84TRR25 pKa = 11.84SGRR28 pKa = 11.84ALINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.79GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 4.8MKK15 pKa = 9.45HH16 pKa = 5.74GFRR19 pKa = 11.84EE20 pKa = 4.06RR21 pKa = 11.84MRR23 pKa = 11.84TRR25 pKa = 11.84SGRR28 pKa = 11.84ALINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.79GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
474780 |
29 |
1774 |
368.9 |
40.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.335 ± 0.074 | 0.748 ± 0.02 |
5.905 ± 0.066 | 5.952 ± 0.068 |
3.519 ± 0.045 | 6.982 ± 0.061 |
2.45 ± 0.033 | 5.083 ± 0.056 |
4.327 ± 0.068 | 9.204 ± 0.071 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.42 ± 0.031 | 3.699 ± 0.047 |
4.604 ± 0.051 | 4.629 ± 0.059 |
5.573 ± 0.068 | 7.195 ± 0.072 |
5.897 ± 0.054 | 7.381 ± 0.061 |
1.406 ± 0.026 | 2.69 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
