
Ruminococcus sp. AM42-11
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Ruminococcus; unclassified Ruminococcus
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
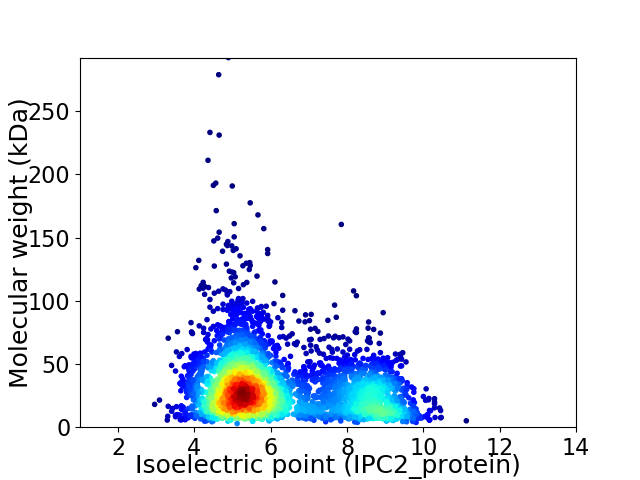
Virtual 2D-PAGE plot for 3917 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A417CV03|A0A417CV03_9FIRM HNH endonuclease OS=Ruminococcus sp. AM42-11 OX=2292372 GN=DW904_10695 PE=4 SV=1
MM1 pKa = 7.41LAGMTLCAVPVMAEE15 pKa = 3.92DD16 pKa = 4.33AEE18 pKa = 4.57TPEE21 pKa = 4.46NEE23 pKa = 4.26VTGDD27 pKa = 3.45ASADD31 pKa = 3.41DD32 pKa = 4.65SFVVWGWNDD41 pKa = 4.05DD42 pKa = 3.25IKK44 pKa = 11.21KK45 pKa = 10.34ILDD48 pKa = 3.8GPFKK52 pKa = 10.32EE53 pKa = 5.41AYY55 pKa = 8.5PDD57 pKa = 3.59DD58 pKa = 3.97YY59 pKa = 11.7EE60 pKa = 6.05RR61 pKa = 11.84IVFVNTGGSDD71 pKa = 3.99YY72 pKa = 10.65YY73 pKa = 10.72QSKK76 pKa = 10.47LDD78 pKa = 4.18PVLDD82 pKa = 4.33DD83 pKa = 4.97PSNEE87 pKa = 4.6LYY89 pKa = 10.16PDD91 pKa = 3.54MMGLEE96 pKa = 4.0VDD98 pKa = 4.13YY99 pKa = 10.89VQKK102 pKa = 10.79YY103 pKa = 8.64VNSDD107 pKa = 2.59WVQNVADD114 pKa = 4.84LGITADD120 pKa = 4.71DD121 pKa = 3.97YY122 pKa = 12.15ANSYY126 pKa = 10.0QYY128 pKa = 11.64NLDD131 pKa = 3.92LGSDD135 pKa = 3.14VDD137 pKa = 4.12GNVRR141 pKa = 11.84ALFWQATPGCYY152 pKa = 9.5AVRR155 pKa = 11.84ADD157 pKa = 4.6LAEE160 pKa = 4.86KK161 pKa = 10.91YY162 pKa = 10.74LGTTDD167 pKa = 3.67PAALAEE173 pKa = 4.31KK174 pKa = 10.35FKK176 pKa = 11.4DD177 pKa = 3.2LDD179 pKa = 4.3TIVATAKK186 pKa = 9.95EE187 pKa = 4.32VNDD190 pKa = 4.27ASGGKK195 pKa = 9.74CKK197 pKa = 10.42LFSGYY202 pKa = 10.71DD203 pKa = 3.42EE204 pKa = 4.74IKK206 pKa = 10.56RR207 pKa = 11.84SLTGSRR213 pKa = 11.84TQGFYY218 pKa = 11.22DD219 pKa = 4.09EE220 pKa = 4.85NDD222 pKa = 3.99KK223 pKa = 9.84ITIDD227 pKa = 4.45DD228 pKa = 4.3NIKK231 pKa = 10.09TYY233 pKa = 10.45MEE235 pKa = 4.22TAKK238 pKa = 9.92TLYY241 pKa = 10.93DD242 pKa = 3.65EE243 pKa = 6.02DD244 pKa = 4.05LTFNTDD250 pKa = 2.19QWSADD255 pKa = 2.68WSANMSGDD263 pKa = 3.64GVDD266 pKa = 3.89SNAALAYY273 pKa = 8.12MGCPWFVYY281 pKa = 9.68WSLSDD286 pKa = 3.06AWKK289 pKa = 10.88GNTILVPTQTKK300 pKa = 9.85CYY302 pKa = 9.28WGGTGLAATTDD313 pKa = 3.87CADD316 pKa = 3.52TEE318 pKa = 4.2LAAKK322 pKa = 9.88IMKK325 pKa = 9.78FFTCDD330 pKa = 3.01EE331 pKa = 4.75DD332 pKa = 4.96GMVAINALNSDD343 pKa = 4.24YY344 pKa = 11.58VNNTAAVSKK353 pKa = 10.13IIEE356 pKa = 4.73AGTSADD362 pKa = 3.52GNGYY366 pKa = 10.13LYY368 pKa = 10.15PDD370 pKa = 4.25AGQNFMEE377 pKa = 5.18FFLPLADD384 pKa = 4.62GLDD387 pKa = 3.5ASMVTAEE394 pKa = 4.37DD395 pKa = 3.72QQILSLMDD403 pKa = 3.66TQTKK407 pKa = 10.37AYY409 pKa = 8.2ATGEE413 pKa = 4.11KK414 pKa = 10.76DD415 pKa = 4.75LDD417 pKa = 3.67TALADD422 pKa = 3.84LTASIHH428 pKa = 6.09DD429 pKa = 3.83TFSYY433 pKa = 10.81LSAEE437 pKa = 4.21
MM1 pKa = 7.41LAGMTLCAVPVMAEE15 pKa = 3.92DD16 pKa = 4.33AEE18 pKa = 4.57TPEE21 pKa = 4.46NEE23 pKa = 4.26VTGDD27 pKa = 3.45ASADD31 pKa = 3.41DD32 pKa = 4.65SFVVWGWNDD41 pKa = 4.05DD42 pKa = 3.25IKK44 pKa = 11.21KK45 pKa = 10.34ILDD48 pKa = 3.8GPFKK52 pKa = 10.32EE53 pKa = 5.41AYY55 pKa = 8.5PDD57 pKa = 3.59DD58 pKa = 3.97YY59 pKa = 11.7EE60 pKa = 6.05RR61 pKa = 11.84IVFVNTGGSDD71 pKa = 3.99YY72 pKa = 10.65YY73 pKa = 10.72QSKK76 pKa = 10.47LDD78 pKa = 4.18PVLDD82 pKa = 4.33DD83 pKa = 4.97PSNEE87 pKa = 4.6LYY89 pKa = 10.16PDD91 pKa = 3.54MMGLEE96 pKa = 4.0VDD98 pKa = 4.13YY99 pKa = 10.89VQKK102 pKa = 10.79YY103 pKa = 8.64VNSDD107 pKa = 2.59WVQNVADD114 pKa = 4.84LGITADD120 pKa = 4.71DD121 pKa = 3.97YY122 pKa = 12.15ANSYY126 pKa = 10.0QYY128 pKa = 11.64NLDD131 pKa = 3.92LGSDD135 pKa = 3.14VDD137 pKa = 4.12GNVRR141 pKa = 11.84ALFWQATPGCYY152 pKa = 9.5AVRR155 pKa = 11.84ADD157 pKa = 4.6LAEE160 pKa = 4.86KK161 pKa = 10.91YY162 pKa = 10.74LGTTDD167 pKa = 3.67PAALAEE173 pKa = 4.31KK174 pKa = 10.35FKK176 pKa = 11.4DD177 pKa = 3.2LDD179 pKa = 4.3TIVATAKK186 pKa = 9.95EE187 pKa = 4.32VNDD190 pKa = 4.27ASGGKK195 pKa = 9.74CKK197 pKa = 10.42LFSGYY202 pKa = 10.71DD203 pKa = 3.42EE204 pKa = 4.74IKK206 pKa = 10.56RR207 pKa = 11.84SLTGSRR213 pKa = 11.84TQGFYY218 pKa = 11.22DD219 pKa = 4.09EE220 pKa = 4.85NDD222 pKa = 3.99KK223 pKa = 9.84ITIDD227 pKa = 4.45DD228 pKa = 4.3NIKK231 pKa = 10.09TYY233 pKa = 10.45MEE235 pKa = 4.22TAKK238 pKa = 9.92TLYY241 pKa = 10.93DD242 pKa = 3.65EE243 pKa = 6.02DD244 pKa = 4.05LTFNTDD250 pKa = 2.19QWSADD255 pKa = 2.68WSANMSGDD263 pKa = 3.64GVDD266 pKa = 3.89SNAALAYY273 pKa = 8.12MGCPWFVYY281 pKa = 9.68WSLSDD286 pKa = 3.06AWKK289 pKa = 10.88GNTILVPTQTKK300 pKa = 9.85CYY302 pKa = 9.28WGGTGLAATTDD313 pKa = 3.87CADD316 pKa = 3.52TEE318 pKa = 4.2LAAKK322 pKa = 9.88IMKK325 pKa = 9.78FFTCDD330 pKa = 3.01EE331 pKa = 4.75DD332 pKa = 4.96GMVAINALNSDD343 pKa = 4.24YY344 pKa = 11.58VNNTAAVSKK353 pKa = 10.13IIEE356 pKa = 4.73AGTSADD362 pKa = 3.52GNGYY366 pKa = 10.13LYY368 pKa = 10.15PDD370 pKa = 4.25AGQNFMEE377 pKa = 5.18FFLPLADD384 pKa = 4.62GLDD387 pKa = 3.5ASMVTAEE394 pKa = 4.37DD395 pKa = 3.72QQILSLMDD403 pKa = 3.66TQTKK407 pKa = 10.37AYY409 pKa = 8.2ATGEE413 pKa = 4.11KK414 pKa = 10.76DD415 pKa = 4.75LDD417 pKa = 3.67TALADD422 pKa = 3.84LTASIHH428 pKa = 6.09DD429 pKa = 3.83TFSYY433 pKa = 10.81LSAEE437 pKa = 4.21
Molecular weight: 47.84 kDa
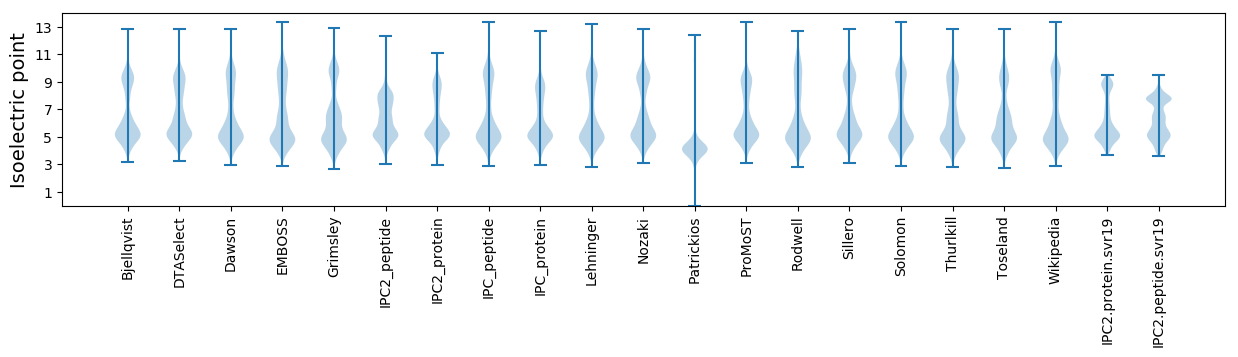
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A417CSJ0|A0A417CSJ0_9FIRM DUF4446 family protein OS=Ruminococcus sp. AM42-11 OX=2292372 GN=DW904_11800 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.15GGRR28 pKa = 11.84KK29 pKa = 8.69VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.68GRR39 pKa = 11.84KK40 pKa = 8.28QLSAA44 pKa = 3.9
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.15GGRR28 pKa = 11.84KK29 pKa = 8.69VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.68GRR39 pKa = 11.84KK40 pKa = 8.28QLSAA44 pKa = 3.9
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1165353 |
24 |
2560 |
297.5 |
33.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.274 ± 0.039 | 1.564 ± 0.02 |
5.74 ± 0.041 | 7.676 ± 0.047 |
4.068 ± 0.025 | 6.989 ± 0.041 |
1.72 ± 0.019 | 7.044 ± 0.031 |
7.22 ± 0.036 | 8.772 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.09 ± 0.022 | 4.524 ± 0.028 |
3.238 ± 0.024 | 3.385 ± 0.024 |
4.2 ± 0.035 | 5.842 ± 0.036 |
5.583 ± 0.046 | 6.778 ± 0.036 |
1.002 ± 0.016 | 4.292 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
