
Betapapillomavirus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Betapapillomavirus
Average proteome isoelectric point is 5.82
Get precalculated fractions of proteins
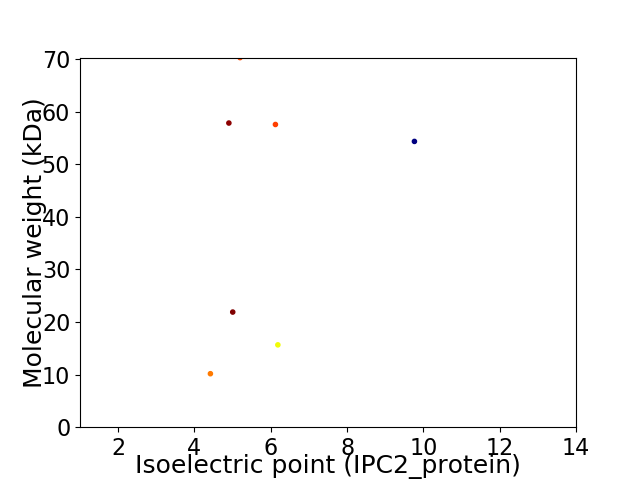
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2AL17|A0A2D2AL17_9PAPI Regulatory protein E2 OS=Betapapillomavirus 4 OX=334208 GN=E2 PE=3 SV=1
MM1 pKa = 7.6IGKK4 pKa = 8.64QATIPEE10 pKa = 4.39IVLDD14 pKa = 4.27LQDD17 pKa = 4.12LVQPIDD23 pKa = 3.44LHH25 pKa = 8.22CDD27 pKa = 3.25EE28 pKa = 5.03EE29 pKa = 4.89LEE31 pKa = 4.28NQVEE35 pKa = 4.37EE36 pKa = 4.62EE37 pKa = 4.23PAPQRR42 pKa = 11.84IDD44 pKa = 3.45YY45 pKa = 10.25KK46 pKa = 10.65IVSSCGGCGIKK57 pKa = 10.43LRR59 pKa = 11.84IFATCTQFGIRR70 pKa = 11.84TLQDD74 pKa = 3.47LLLEE78 pKa = 4.9EE79 pKa = 4.78IALLCPDD86 pKa = 4.81CKK88 pKa = 10.68NGRR91 pKa = 3.72
MM1 pKa = 7.6IGKK4 pKa = 8.64QATIPEE10 pKa = 4.39IVLDD14 pKa = 4.27LQDD17 pKa = 4.12LVQPIDD23 pKa = 3.44LHH25 pKa = 8.22CDD27 pKa = 3.25EE28 pKa = 5.03EE29 pKa = 4.89LEE31 pKa = 4.28NQVEE35 pKa = 4.37EE36 pKa = 4.62EE37 pKa = 4.23PAPQRR42 pKa = 11.84IDD44 pKa = 3.45YY45 pKa = 10.25KK46 pKa = 10.65IVSSCGGCGIKK57 pKa = 10.43LRR59 pKa = 11.84IFATCTQFGIRR70 pKa = 11.84TLQDD74 pKa = 3.47LLLEE78 pKa = 4.9EE79 pKa = 4.78IALLCPDD86 pKa = 4.81CKK88 pKa = 10.68NGRR91 pKa = 3.72
Molecular weight: 10.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2AL52|A0A2D2AL52_9PAPI Protein E6 OS=Betapapillomavirus 4 OX=334208 GN=E6 PE=3 SV=1
MM1 pKa = 7.5EE2 pKa = 5.42ALSDD6 pKa = 4.12RR7 pKa = 11.84FNALQDD13 pKa = 3.51KK14 pKa = 10.97LMTIYY19 pKa = 10.57EE20 pKa = 4.33KK21 pKa = 10.81ASEE24 pKa = 4.26SLTDD28 pKa = 5.54QIEE31 pKa = 4.11HH32 pKa = 6.5WNLLRR37 pKa = 11.84QEE39 pKa = 4.21QVLFHH44 pKa = 6.04YY45 pKa = 10.25ARR47 pKa = 11.84QRR49 pKa = 11.84GVMRR53 pKa = 11.84LGYY56 pKa = 10.1QPVPALTISEE66 pKa = 4.36AKK68 pKa = 10.21AKK70 pKa = 10.0EE71 pKa = 4.12AIAMVLHH78 pKa = 6.59LEE80 pKa = 4.11ALQRR84 pKa = 11.84SPYY87 pKa = 10.01KK88 pKa = 10.23DD89 pKa = 3.66EE90 pKa = 4.24KK91 pKa = 8.2WTLVNTSVEE100 pKa = 4.39TFRR103 pKa = 11.84TPPEE107 pKa = 3.76NCFKK111 pKa = 10.82KK112 pKa = 10.56GPKK115 pKa = 9.09TIEE118 pKa = 3.58IVYY121 pKa = 10.4DD122 pKa = 3.78GNPDD126 pKa = 2.96NTMLYY131 pKa = 9.05TIWTFIYY138 pKa = 11.02YY139 pKa = 10.31EE140 pKa = 4.44DD141 pKa = 6.11DD142 pKa = 3.26EE143 pKa = 5.95GNWQKK148 pKa = 9.59TEE150 pKa = 3.87GQLDD154 pKa = 3.69YY155 pKa = 11.36HH156 pKa = 6.29GAYY159 pKa = 10.5YY160 pKa = 10.95LDD162 pKa = 4.19GLNKK166 pKa = 9.71QYY168 pKa = 10.88YY169 pKa = 9.58IRR171 pKa = 11.84FAQDD175 pKa = 2.32ARR177 pKa = 11.84RR178 pKa = 11.84YY179 pKa = 9.75SEE181 pKa = 3.82TGEE184 pKa = 4.01WEE186 pKa = 4.2VKK188 pKa = 10.11FNNEE192 pKa = 3.3ILFAPVTSSTNSEE205 pKa = 4.13EE206 pKa = 4.22EE207 pKa = 3.85RR208 pKa = 11.84DD209 pKa = 3.94RR210 pKa = 11.84PAPASDD216 pKa = 4.23PGSLPQTSGRR226 pKa = 11.84LSPSPVQRR234 pKa = 11.84KK235 pKa = 7.21QLSKK239 pKa = 10.48RR240 pKa = 11.84RR241 pKa = 11.84YY242 pKa = 7.71GRR244 pKa = 11.84KK245 pKa = 9.65DD246 pKa = 3.22SSPTATSRR254 pKa = 11.84VQRR257 pKa = 11.84RR258 pKa = 11.84PKK260 pKa = 10.18ASPRR264 pKa = 11.84RR265 pKa = 11.84SRR267 pKa = 11.84SRR269 pKa = 11.84SGSRR273 pKa = 11.84SGSQGDD279 pKa = 3.62ARR281 pKa = 11.84TLLSLRR287 pKa = 11.84RR288 pKa = 11.84GEE290 pKa = 4.1RR291 pKa = 11.84EE292 pKa = 3.63RR293 pKa = 11.84GEE295 pKa = 4.04GRR297 pKa = 11.84GTGIRR302 pKa = 11.84VRR304 pKa = 11.84GRR306 pKa = 11.84SGSRR310 pKa = 11.84GRR312 pKa = 11.84SRR314 pKa = 11.84SRR316 pKa = 11.84SRR318 pKa = 11.84SRR320 pKa = 11.84SRR322 pKa = 11.84SRR324 pKa = 11.84SRR326 pKa = 11.84SRR328 pKa = 11.84SSRR331 pKa = 11.84SRR333 pKa = 11.84GRR335 pKa = 11.84GNTTTRR341 pKa = 11.84GRR343 pKa = 11.84GGYY346 pKa = 7.01RR347 pKa = 11.84TRR349 pKa = 11.84RR350 pKa = 11.84SRR352 pKa = 11.84SKK354 pKa = 11.01SVDD357 pKa = 3.07TSGIPPDD364 pKa = 3.38QVGRR368 pKa = 11.84SLQSVGRR375 pKa = 11.84QHH377 pKa = 6.97SGRR380 pKa = 11.84LARR383 pKa = 11.84LLDD386 pKa = 3.84EE387 pKa = 5.59ARR389 pKa = 11.84DD390 pKa = 3.77PPVILLKK397 pKa = 11.05GKK399 pKa = 10.59ANTLKK404 pKa = 10.49CYY406 pKa = 10.29RR407 pKa = 11.84YY408 pKa = 9.45RR409 pKa = 11.84AKK411 pKa = 10.46EE412 pKa = 3.99KK413 pKa = 10.72YY414 pKa = 8.94KK415 pKa = 10.68GYY417 pKa = 9.79YY418 pKa = 10.1DD419 pKa = 3.54RR420 pKa = 11.84VSTTWSWVGAGSNDD434 pKa = 3.64RR435 pKa = 11.84IGRR438 pKa = 11.84SRR440 pKa = 11.84MLFSFTSKK448 pKa = 9.95SQRR451 pKa = 11.84QMFLNIMKK459 pKa = 10.12LPKK462 pKa = 10.5GVDD465 pKa = 2.93WSFGCFDD472 pKa = 4.93SII474 pKa = 5.05
MM1 pKa = 7.5EE2 pKa = 5.42ALSDD6 pKa = 4.12RR7 pKa = 11.84FNALQDD13 pKa = 3.51KK14 pKa = 10.97LMTIYY19 pKa = 10.57EE20 pKa = 4.33KK21 pKa = 10.81ASEE24 pKa = 4.26SLTDD28 pKa = 5.54QIEE31 pKa = 4.11HH32 pKa = 6.5WNLLRR37 pKa = 11.84QEE39 pKa = 4.21QVLFHH44 pKa = 6.04YY45 pKa = 10.25ARR47 pKa = 11.84QRR49 pKa = 11.84GVMRR53 pKa = 11.84LGYY56 pKa = 10.1QPVPALTISEE66 pKa = 4.36AKK68 pKa = 10.21AKK70 pKa = 10.0EE71 pKa = 4.12AIAMVLHH78 pKa = 6.59LEE80 pKa = 4.11ALQRR84 pKa = 11.84SPYY87 pKa = 10.01KK88 pKa = 10.23DD89 pKa = 3.66EE90 pKa = 4.24KK91 pKa = 8.2WTLVNTSVEE100 pKa = 4.39TFRR103 pKa = 11.84TPPEE107 pKa = 3.76NCFKK111 pKa = 10.82KK112 pKa = 10.56GPKK115 pKa = 9.09TIEE118 pKa = 3.58IVYY121 pKa = 10.4DD122 pKa = 3.78GNPDD126 pKa = 2.96NTMLYY131 pKa = 9.05TIWTFIYY138 pKa = 11.02YY139 pKa = 10.31EE140 pKa = 4.44DD141 pKa = 6.11DD142 pKa = 3.26EE143 pKa = 5.95GNWQKK148 pKa = 9.59TEE150 pKa = 3.87GQLDD154 pKa = 3.69YY155 pKa = 11.36HH156 pKa = 6.29GAYY159 pKa = 10.5YY160 pKa = 10.95LDD162 pKa = 4.19GLNKK166 pKa = 9.71QYY168 pKa = 10.88YY169 pKa = 9.58IRR171 pKa = 11.84FAQDD175 pKa = 2.32ARR177 pKa = 11.84RR178 pKa = 11.84YY179 pKa = 9.75SEE181 pKa = 3.82TGEE184 pKa = 4.01WEE186 pKa = 4.2VKK188 pKa = 10.11FNNEE192 pKa = 3.3ILFAPVTSSTNSEE205 pKa = 4.13EE206 pKa = 4.22EE207 pKa = 3.85RR208 pKa = 11.84DD209 pKa = 3.94RR210 pKa = 11.84PAPASDD216 pKa = 4.23PGSLPQTSGRR226 pKa = 11.84LSPSPVQRR234 pKa = 11.84KK235 pKa = 7.21QLSKK239 pKa = 10.48RR240 pKa = 11.84RR241 pKa = 11.84YY242 pKa = 7.71GRR244 pKa = 11.84KK245 pKa = 9.65DD246 pKa = 3.22SSPTATSRR254 pKa = 11.84VQRR257 pKa = 11.84RR258 pKa = 11.84PKK260 pKa = 10.18ASPRR264 pKa = 11.84RR265 pKa = 11.84SRR267 pKa = 11.84SRR269 pKa = 11.84SGSRR273 pKa = 11.84SGSQGDD279 pKa = 3.62ARR281 pKa = 11.84TLLSLRR287 pKa = 11.84RR288 pKa = 11.84GEE290 pKa = 4.1RR291 pKa = 11.84EE292 pKa = 3.63RR293 pKa = 11.84GEE295 pKa = 4.04GRR297 pKa = 11.84GTGIRR302 pKa = 11.84VRR304 pKa = 11.84GRR306 pKa = 11.84SGSRR310 pKa = 11.84GRR312 pKa = 11.84SRR314 pKa = 11.84SRR316 pKa = 11.84SRR318 pKa = 11.84SRR320 pKa = 11.84SRR322 pKa = 11.84SRR324 pKa = 11.84SRR326 pKa = 11.84SRR328 pKa = 11.84SSRR331 pKa = 11.84SRR333 pKa = 11.84GRR335 pKa = 11.84GNTTTRR341 pKa = 11.84GRR343 pKa = 11.84GGYY346 pKa = 7.01RR347 pKa = 11.84TRR349 pKa = 11.84RR350 pKa = 11.84SRR352 pKa = 11.84SKK354 pKa = 11.01SVDD357 pKa = 3.07TSGIPPDD364 pKa = 3.38QVGRR368 pKa = 11.84SLQSVGRR375 pKa = 11.84QHH377 pKa = 6.97SGRR380 pKa = 11.84LARR383 pKa = 11.84LLDD386 pKa = 3.84EE387 pKa = 5.59ARR389 pKa = 11.84DD390 pKa = 3.77PPVILLKK397 pKa = 11.05GKK399 pKa = 10.59ANTLKK404 pKa = 10.49CYY406 pKa = 10.29RR407 pKa = 11.84YY408 pKa = 9.45RR409 pKa = 11.84AKK411 pKa = 10.46EE412 pKa = 3.99KK413 pKa = 10.72YY414 pKa = 8.94KK415 pKa = 10.68GYY417 pKa = 9.79YY418 pKa = 10.1DD419 pKa = 3.54RR420 pKa = 11.84VSTTWSWVGAGSNDD434 pKa = 3.64RR435 pKa = 11.84IGRR438 pKa = 11.84SRR440 pKa = 11.84MLFSFTSKK448 pKa = 9.95SQRR451 pKa = 11.84QMFLNIMKK459 pKa = 10.12LPKK462 pKa = 10.5GVDD465 pKa = 2.93WSFGCFDD472 pKa = 4.93SII474 pKa = 5.05
Molecular weight: 54.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2554 |
91 |
612 |
364.9 |
41.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.951 ± 0.4 | 2.271 ± 0.86 |
6.46 ± 0.427 | 6.852 ± 0.545 |
3.876 ± 0.425 | 6.891 ± 0.792 |
1.879 ± 0.257 | 4.777 ± 0.654 |
5.442 ± 0.789 | 8.614 ± 0.78 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.331 ± 0.281 | 4.15 ± 0.551 |
5.873 ± 0.778 | 4.62 ± 0.282 |
7.009 ± 1.4 | 7.244 ± 1.023 |
6.147 ± 0.908 | 5.912 ± 0.679 |
1.253 ± 0.292 | 3.446 ± 0.246 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
